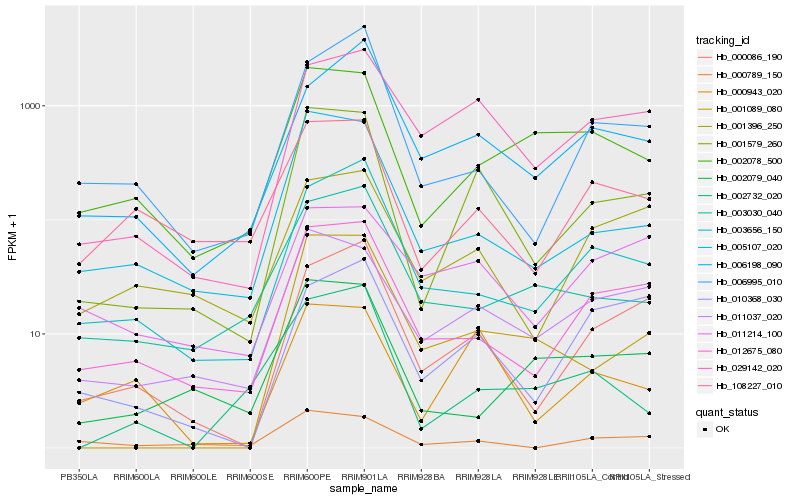
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001579_260 |
0.0 |
- |
- |
putative ADP-ribolylation factor [Arabidopsis thaliana] |
| 2 |
Hb_011037_020 |
0.171744672 |
- |
- |
unknown [Picea sitchensis] |
| 3 |
Hb_005107_020 |
0.1728363794 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 4 |
Hb_029142_020 |
0.1856324175 |
- |
- |
hypothetical protein POPTR_0004s23900g [Populus trichocarpa] |
| 5 |
Hb_012675_080 |
0.1862779823 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 6 |
Hb_000086_190 |
0.1869228746 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 3-like protein [Cucumis sativus] |
| 7 |
Hb_000943_020 |
0.1869339474 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 8 |
Hb_001089_080 |
0.2009155887 |
- |
- |
hypothetical protein glysoja_044047 [Glycine soja] |
| 9 |
Hb_000789_150 |
0.2119613223 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_108227_010 |
0.2127546758 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 11 |
Hb_002078_500 |
0.2147643581 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003656_150 |
0.2154732178 |
- |
- |
- |
| 13 |
Hb_001396_250 |
0.2280770477 |
- |
- |
PREDICTED: stress-associated endoplasmic reticulum protein 2 [Jatropha curcas] |
| 14 |
Hb_006995_010 |
0.2300490419 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein [Jatropha curcas] |
| 15 |
Hb_006198_090 |
0.2332440437 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |
| 16 |
Hb_010368_030 |
0.2349491955 |
- |
- |
BnaA07g35570D [Brassica napus] |
| 17 |
Hb_003030_040 |
0.2374233966 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 18 |
Hb_002732_020 |
0.2389861518 |
- |
- |
- |
| 19 |
Hb_011214_100 |
0.2403494065 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 20 |
Hb_002079_040 |
0.2409209347 |
- |
- |
hypothetical protein VITISV_018542 [Vitis vinifera] |