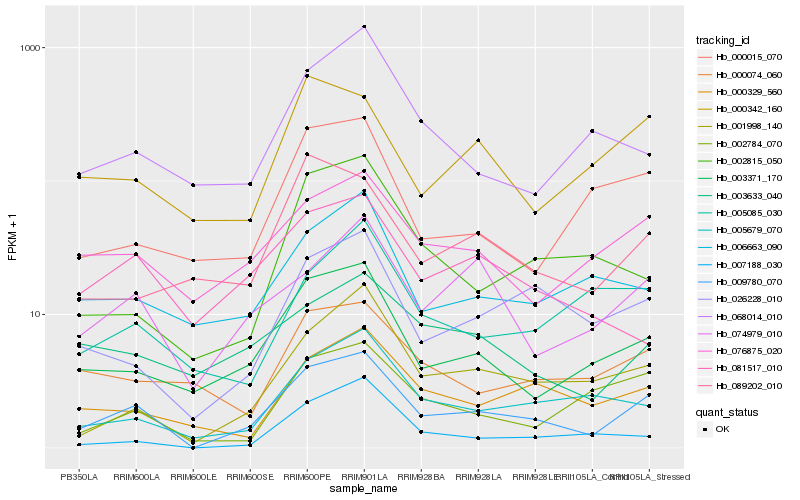
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009780_070 |
0.0 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1 [Jatropha curcas] |
| 2 |
Hb_003371_170 |
0.1775710961 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001998_140 |
0.1933854823 |
- |
- |
- |
| 4 |
Hb_076875_020 |
0.1953099006 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 5 |
Hb_026228_010 |
0.1961597206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_074979_010 |
0.1983626773 |
- |
- |
hypothetical protein AALP_AA2G213100 [Arabis alpina] |
| 7 |
Hb_005679_070 |
0.2023754484 |
- |
- |
hypothetical protein M569_07741, partial [Genlisea aurea] |
| 8 |
Hb_000329_560 |
0.2031544681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002784_070 |
0.2098731817 |
- |
- |
BnaUnng01470D [Brassica napus] |
| 10 |
Hb_006663_090 |
0.2149879802 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 11 |
Hb_000342_160 |
0.2155919468 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 12 |
Hb_003633_040 |
0.2180033065 |
- |
- |
hypothetical protein B456_008G146200 [Gossypium raimondii] |
| 13 |
Hb_000074_060 |
0.2188093028 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 14 |
Hb_089202_010 |
0.2196705565 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Nicotiana sylvestris] |
| 15 |
Hb_000015_070 |
0.2197065996 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |
| 16 |
Hb_005085_030 |
0.227813068 |
- |
- |
PREDICTED: N-acetyltransferase 9-like protein isoform X1 [Jatropha curcas] |
| 17 |
Hb_081517_010 |
0.2294032499 |
- |
- |
PREDICTED: putative potassium transporter 12 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002815_050 |
0.2303632284 |
- |
- |
PREDICTED: succinate-semialdehyde dehydrogenase, mitochondrial [Populus euphratica] |
| 19 |
Hb_007188_030 |
0.2310067301 |
- |
- |
PREDICTED: DNA topoisomerase 3-alpha-like [Malus domestica] |
| 20 |
Hb_068014_010 |
0.2324727253 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |