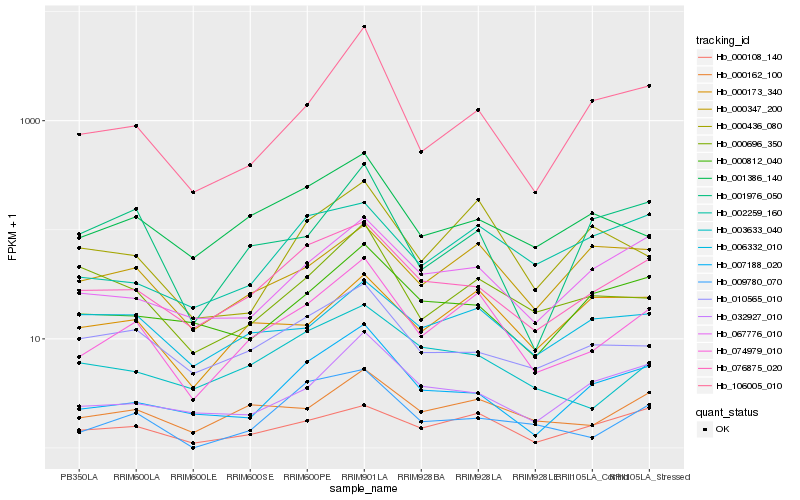
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074979_010 |
0.0 |
- |
- |
hypothetical protein AALP_AA2G213100 [Arabis alpina] |
| 2 |
Hb_000162_100 |
0.1386607504 |
- |
- |
PREDICTED: uncharacterized protein LOC105638832 [Jatropha curcas] |
| 3 |
Hb_076875_020 |
0.1599322246 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 4 |
Hb_000108_140 |
0.1676465142 |
- |
- |
hypothetical protein CICLE_v10019690mg [Citrus clementina] |
| 5 |
Hb_003633_040 |
0.1820889483 |
- |
- |
hypothetical protein B456_008G146200 [Gossypium raimondii] |
| 6 |
Hb_067776_010 |
0.1822065951 |
- |
- |
mitochondrial 39S ribosomal protein L53 [Hevea brasiliensis] |
| 7 |
Hb_001386_140 |
0.1825160294 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 8 |
Hb_000436_080 |
0.1825805574 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_000347_200 |
0.188310856 |
- |
- |
lsm1, putative [Ricinus communis] |
| 10 |
Hb_007188_020 |
0.1892593595 |
- |
- |
hypothetical protein PRUPE_ppb010436mg [Prunus persica] |
| 11 |
Hb_106005_010 |
0.190794662 |
- |
- |
EG5651 [Manihot esculenta] |
| 12 |
Hb_000173_340 |
0.1925524463 |
- |
- |
hypothetical protein JCGZ_02075 [Jatropha curcas] |
| 13 |
Hb_010565_010 |
0.1927790756 |
- |
- |
PREDICTED: N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase-like isoform X2 [Populus euphratica] |
| 14 |
Hb_000696_350 |
0.1932975599 |
- |
- |
ARM repeat superfamily protein isoform 2 [Theobroma cacao] |
| 15 |
Hb_032927_010 |
0.1952406973 |
- |
- |
- |
| 16 |
Hb_000812_040 |
0.1979256703 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 17 |
Hb_001976_050 |
0.1981117047 |
- |
- |
PREDICTED: protein SKIP34 [Jatropha curcas] |
| 18 |
Hb_009780_070 |
0.1983626773 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1 [Jatropha curcas] |
| 19 |
Hb_006332_010 |
0.198706706 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 isoform X1 [Vitis vinifera] |
| 20 |
Hb_002259_160 |
0.1992418457 |
- |
- |
hypothetical protein PSEUBRA_SCAF14g01508 [Pseudozyma brasiliensis GHG001] |