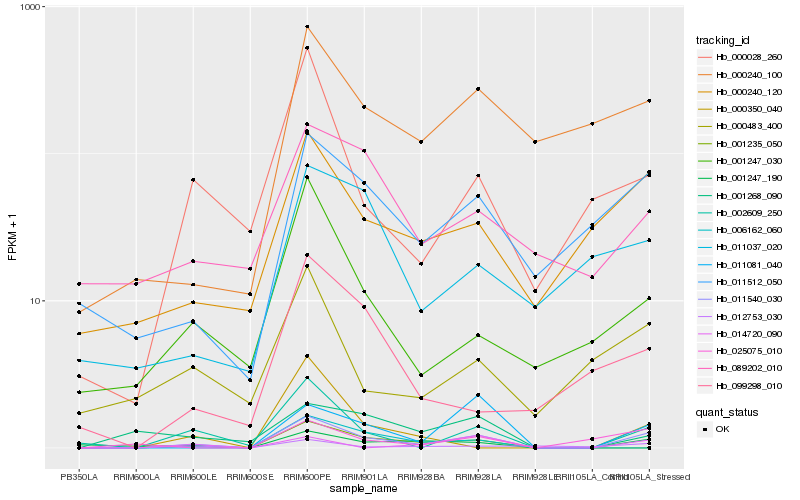
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011540_030 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX5-like isoform X6 [Glycine max] |
| 2 |
Hb_002609_250 |
0.2476703164 |
- |
- |
PREDICTED: starch-binding domain-containing protein 1-like [Jatropha curcas] |
| 3 |
Hb_001247_190 |
0.2717147512 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_011081_040 |
0.2774983636 |
- |
- |
hypothetical protein JCGZ_20956 [Jatropha curcas] |
| 5 |
Hb_011512_050 |
0.2972593123 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 13-A isoform 2 [Theobroma cacao] |
| 6 |
Hb_000240_120 |
0.3018802078 |
- |
- |
ras-related protein Rab-5B [Reticulomyxa filosa] |
| 7 |
Hb_000028_260 |
0.3174951769 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 8 |
Hb_025075_010 |
0.3176369751 |
- |
- |
50S ribosomal protein L24 [Glycine soja] |
| 9 |
Hb_012753_030 |
0.3187846692 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |
| 10 |
Hb_000240_100 |
0.3196000869 |
- |
- |
PREDICTED: ras-related protein Rab5-like [Eucalyptus grandis] |
| 11 |
Hb_001247_030 |
0.3253407718 |
- |
- |
- |
| 12 |
Hb_000350_040 |
0.3301140595 |
- |
- |
- |
| 13 |
Hb_014720_090 |
0.3303030535 |
- |
- |
- |
| 14 |
Hb_001268_090 |
0.3308045302 |
- |
- |
Uncharacterized protein TCM_020504 [Theobroma cacao] |
| 15 |
Hb_001235_050 |
0.3335652542 |
- |
- |
PREDICTED: proteasome activator subunit 4 [Nelumbo nucifera] |
| 16 |
Hb_000483_400 |
0.3408466976 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_099298_010 |
0.3422961706 |
- |
- |
- |
| 18 |
Hb_089202_010 |
0.3432322967 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Nicotiana sylvestris] |
| 19 |
Hb_011037_020 |
0.3442483739 |
- |
- |
unknown [Picea sitchensis] |
| 20 |
Hb_006162_060 |
0.344409034 |
- |
- |
PREDICTED: LYR motif-containing protein At3g19508 [Jatropha curcas] |