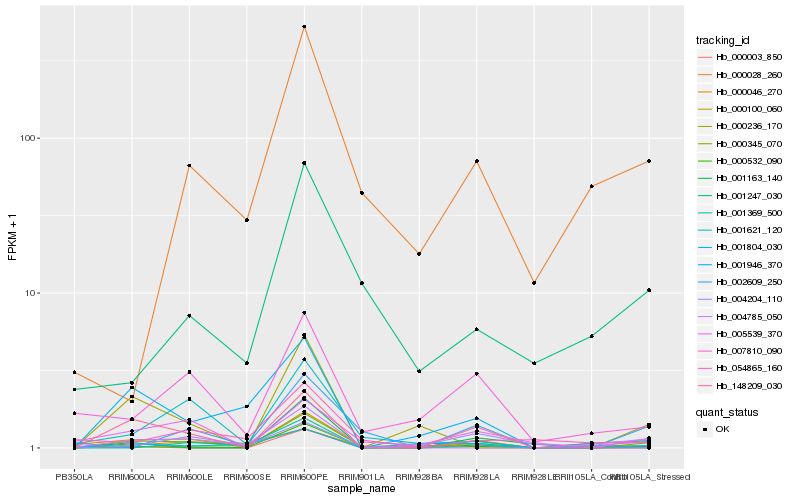
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_070 |
0.0 |
rubber biosynthesis |
Gene Name: SRPP6 |
- |
| 2 |
Hb_000532_090 |
0.2531339226 |
- |
- |
ATPase [delta proteobacterium MLMS-1] |
| 3 |
Hb_000100_060 |
0.2674511187 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Populus euphratica] |
| 4 |
Hb_148209_030 |
0.2734397184 |
- |
- |
hypothetical protein VITISV_017120 [Vitis vinifera] |
| 5 |
Hb_004785_050 |
0.2817561385 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 6 |
Hb_002609_250 |
0.2831699732 |
- |
- |
PREDICTED: starch-binding domain-containing protein 1-like [Jatropha curcas] |
| 7 |
Hb_001621_120 |
0.2907051034 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 8 |
Hb_005539_370 |
0.2925191972 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 9 |
Hb_001804_030 |
0.2969766809 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_007810_090 |
0.3003339706 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |
| 11 |
Hb_000046_270 |
0.3106276812 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000028_260 |
0.3133285268 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 13 |
Hb_001369_500 |
0.3170191048 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 14 |
Hb_000003_850 |
0.3174629204 |
- |
- |
PREDICTED: uncharacterized protein LOC105633597 [Jatropha curcas] |
| 15 |
Hb_001946_370 |
0.3185091827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001247_030 |
0.3210374974 |
- |
- |
- |
| 17 |
Hb_001163_140 |
0.3215914012 |
- |
- |
- |
| 18 |
Hb_054865_160 |
0.3217953611 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 19 |
Hb_000236_170 |
0.3238924448 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 20 |
Hb_004204_110 |
0.3243841085 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |