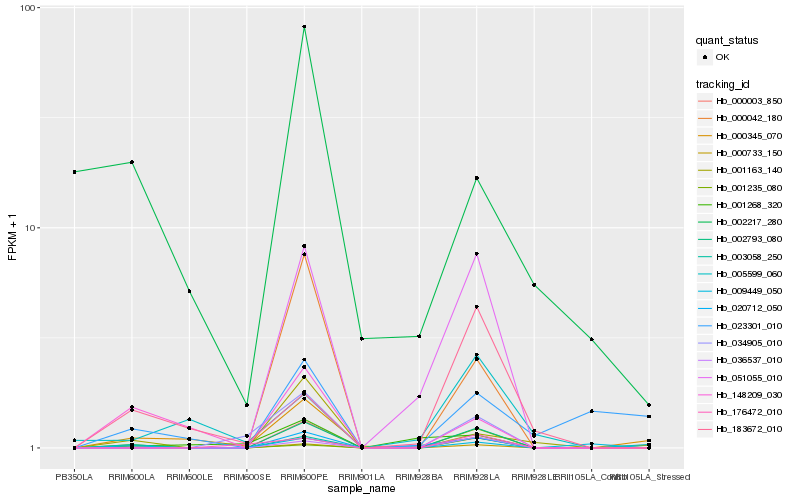
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003058_250 |
0.0 |
- |
- |
hypothetical protein RCOM_0006440 [Ricinus communis] |
| 2 |
Hb_000733_150 |
0.2587148313 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Populus euphratica] |
| 3 |
Hb_002793_080 |
0.2598062731 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 4 |
Hb_036537_010 |
0.2836800597 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000003_850 |
0.2941508139 |
- |
- |
PREDICTED: uncharacterized protein LOC105633597 [Jatropha curcas] |
| 6 |
Hb_051055_010 |
0.2979810741 |
- |
- |
- |
| 7 |
Hb_009449_050 |
0.3206265005 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Nelumbo nucifera] |
| 8 |
Hb_148209_030 |
0.3228422893 |
- |
- |
hypothetical protein VITISV_017120 [Vitis vinifera] |
| 9 |
Hb_020712_050 |
0.3295396194 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 10 |
Hb_001235_080 |
0.3476152863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_034905_010 |
0.3559162709 |
- |
- |
PREDICTED: uncharacterized protein LOC104891359 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_000042_180 |
0.3565084611 |
- |
- |
- |
| 13 |
Hb_001163_140 |
0.363257282 |
- |
- |
- |
| 14 |
Hb_183672_010 |
0.3659856972 |
- |
- |
PREDICTED: (S)-tetrahydroprotoberberine N-methyltransferase isoform X3 [Jatropha curcas] |
| 15 |
Hb_176472_010 |
0.3722809308 |
- |
- |
BnaC01g33060D [Brassica napus] |
| 16 |
Hb_005599_060 |
0.4199615444 |
- |
- |
PREDICTED: syntaxin-72 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000345_070 |
0.4211954748 |
rubber biosynthesis |
Gene Name: SRPP6 |
- |
| 18 |
Hb_001268_320 |
0.4247302228 |
- |
- |
PREDICTED: uncharacterized protein LOC104887816 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_002217_280 |
0.4267424296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_023301_010 |
0.431349867 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |