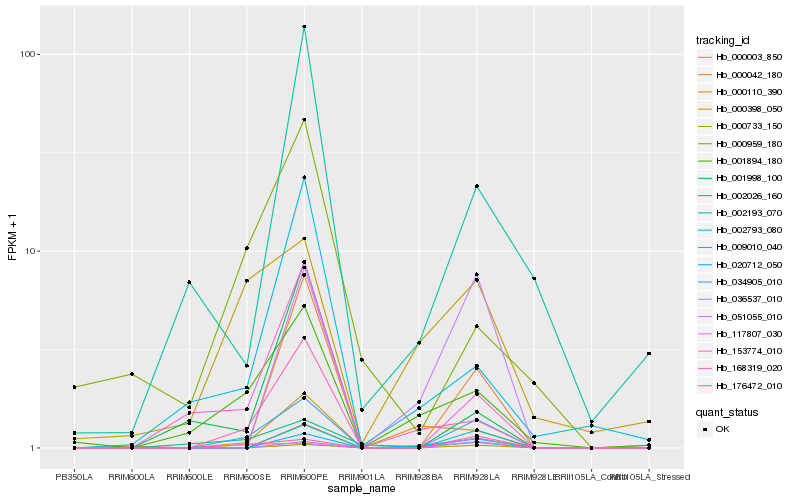
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034905_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104891359 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_176472_010 |
0.2056587916 |
- |
- |
BnaC01g33060D [Brassica napus] |
| 3 |
Hb_002793_080 |
0.2433375175 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 4 |
Hb_000733_150 |
0.2464869345 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Populus euphratica] |
| 5 |
Hb_117807_030 |
0.2465131144 |
- |
- |
Protein MCM10 [Gossypium arboreum] |
| 6 |
Hb_000042_180 |
0.26615814 |
- |
- |
- |
| 7 |
Hb_168319_020 |
0.2735428117 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_153774_010 |
0.2868994043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001894_180 |
0.2871425156 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 10 |
Hb_051055_010 |
0.3011131197 |
- |
- |
- |
| 11 |
Hb_020712_050 |
0.3036536388 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 12 |
Hb_036537_010 |
0.3284683396 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_000398_050 |
0.3348125276 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 14 |
Hb_002026_160 |
0.3381449474 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-like [Jatropha curcas] |
| 15 |
Hb_000003_850 |
0.3456876121 |
- |
- |
PREDICTED: uncharacterized protein LOC105633597 [Jatropha curcas] |
| 16 |
Hb_002193_070 |
0.3483887967 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_001998_100 |
0.3484937863 |
- |
- |
- |
| 18 |
Hb_009010_040 |
0.3493621925 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 19 |
Hb_000110_390 |
0.3533873416 |
- |
- |
PREDICTED: uncharacterized protein LOC103338992 [Prunus mume] |
| 20 |
Hb_000959_180 |
0.3537660932 |
- |
- |
hypothetical protein JCGZ_09448 [Jatropha curcas] |