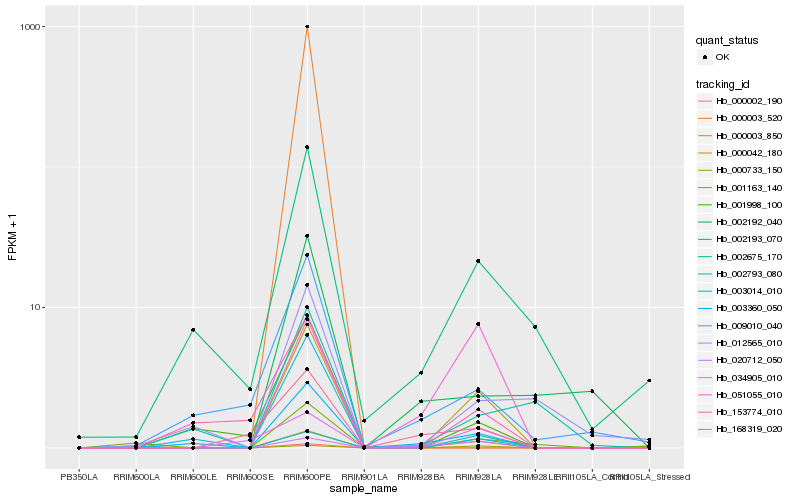
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_180 |
0.0 |
- |
- |
- |
| 2 |
Hb_003360_050 |
0.1974008574 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 3 |
Hb_002793_080 |
0.2187437789 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 4 |
Hb_000733_150 |
0.227966929 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Populus euphratica] |
| 5 |
Hb_001998_100 |
0.2443129819 |
- |
- |
- |
| 6 |
Hb_002193_070 |
0.249300907 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_003014_010 |
0.255126995 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0017s11800g [Populus trichocarpa] |
| 8 |
Hb_001163_140 |
0.260081428 |
- |
- |
- |
| 9 |
Hb_153774_010 |
0.2611266896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_034905_010 |
0.26615814 |
- |
- |
PREDICTED: uncharacterized protein LOC104891359 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_020712_050 |
0.2677095031 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 12 |
Hb_000003_850 |
0.272867981 |
- |
- |
PREDICTED: uncharacterized protein LOC105633597 [Jatropha curcas] |
| 13 |
Hb_009010_040 |
0.2820479157 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 14 |
Hb_168319_020 |
0.2821257881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_012565_010 |
0.2895847915 |
- |
- |
PREDICTED: putative glutamine amidotransferase PB2B2.05 [Jatropha curcas] |
| 16 |
Hb_051055_010 |
0.3036273497 |
- |
- |
- |
| 17 |
Hb_002675_170 |
0.3084892574 |
- |
- |
hypothetical protein JCGZ_06235 [Jatropha curcas] |
| 18 |
Hb_002192_040 |
0.3104862898 |
- |
- |
- |
| 19 |
Hb_000002_190 |
0.3191427447 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor [Morus notabilis] |
| 20 |
Hb_000003_520 |
0.3191427447 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |