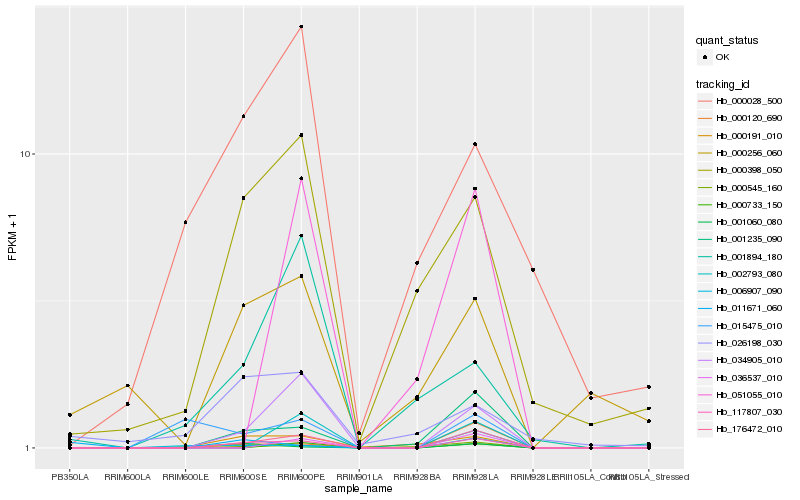
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_117807_030 |
0.0 |
- |
- |
Protein MCM10 [Gossypium arboreum] |
| 2 |
Hb_176472_010 |
0.1166398416 |
- |
- |
BnaC01g33060D [Brassica napus] |
| 3 |
Hb_001235_090 |
0.2134846288 |
- |
- |
PREDICTED: uncharacterized protein LOC103449952 [Malus domestica] |
| 4 |
Hb_034905_010 |
0.2465131144 |
- |
- |
PREDICTED: uncharacterized protein LOC104891359 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_000191_010 |
0.2745634119 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 6 |
Hb_000545_160 |
0.2779870033 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 7 |
Hb_000120_690 |
0.3057334879 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 8 |
Hb_000398_050 |
0.319133041 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 9 |
Hb_006907_090 |
0.3292495827 |
- |
- |
- |
| 10 |
Hb_001060_080 |
0.3327810406 |
- |
- |
PREDICTED: uncharacterized protein LOC104219776 [Nicotiana sylvestris] |
| 11 |
Hb_036537_010 |
0.3657009999 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000256_060 |
0.3770023111 |
- |
- |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 13 |
Hb_000733_150 |
0.3800234429 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Populus euphratica] |
| 14 |
Hb_002793_080 |
0.3825963154 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 15 |
Hb_011671_060 |
0.3852419663 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 16 |
Hb_026198_030 |
0.3900085031 |
- |
- |
PREDICTED: ABC transporter G family member 25 [Jatropha curcas] |
| 17 |
Hb_015475_010 |
0.3973290086 |
- |
- |
hypothetical protein VITISV_034342 [Vitis vinifera] |
| 18 |
Hb_000028_500 |
0.3978897992 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_051055_010 |
0.3981297198 |
- |
- |
- |
| 20 |
Hb_001894_180 |
0.4028848681 |
- |
- |
galactinol synthase [Manihot esculenta] |