| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
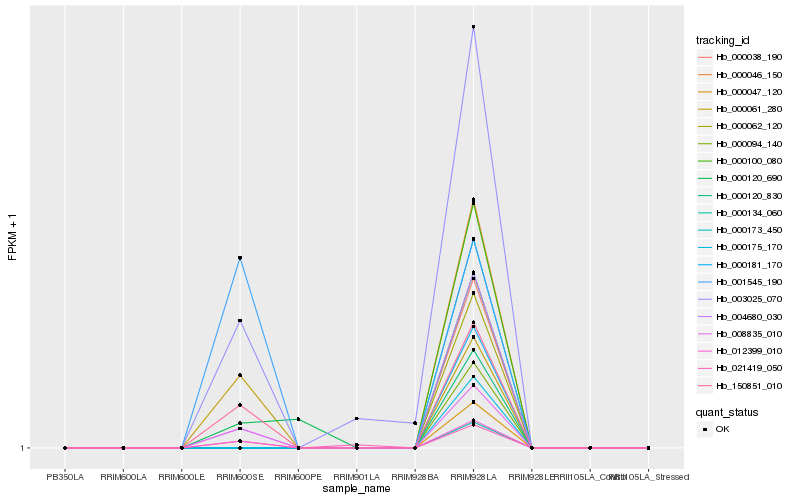
Hb_012399_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_008835_010 |
0.0382578461 |
- |
- |
PREDICTED: uncharacterized protein LOC105851721 [Cicer arietinum] |
| 3 |
Hb_150851_010 |
0.0512742026 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_004680_030 |
0.1270469082 |
- |
- |
PREDICTED: uncharacterized protein LOC105795864 [Gossypium raimondii] |
| 5 |
Hb_000061_280 |
0.1828449943 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g44230 [Jatropha curcas] |
| 6 |
Hb_003025_070 |
0.1962695855 |
- |
- |
PREDICTED: uncharacterized protein LOC103419743 [Malus domestica] |
| 7 |
Hb_021419_050 |
0.2210998892 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Malus domestica] |
| 8 |
Hb_001545_190 |
0.2534399609 |
- |
- |
hypothetical protein VITISV_012031 [Vitis vinifera] |
| 9 |
Hb_000120_690 |
0.2647187554 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 10 |
Hb_000038_190 |
0.3247443005 |
- |
- |
- |
| 11 |
Hb_000046_150 |
0.3247443005 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000047_120 |
0.3247443005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000062_120 |
0.3247443005 |
- |
- |
hypothetical protein JCGZ_17895 [Jatropha curcas] |
| 14 |
Hb_000094_140 |
0.3247443005 |
- |
- |
hypothetical protein POPTR_0007s09865g [Populus trichocarpa] |
| 15 |
Hb_000100_080 |
0.3247443005 |
- |
- |
- |
| 16 |
Hb_000120_830 |
0.3247443005 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 17 |
Hb_000134_060 |
0.3247443005 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: extended synaptotagmin-1-like [Jatropha curcas] |
| 18 |
Hb_000173_450 |
0.3247443005 |
- |
- |
- |
| 19 |
Hb_000175_170 |
0.3247443005 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit gamma-like PV42a [Jatropha curcas] |
| 20 |
Hb_000181_170 |
0.3247443005 |
- |
- |
hypothetical protein POPTR_0011s11750g [Populus trichocarpa] |