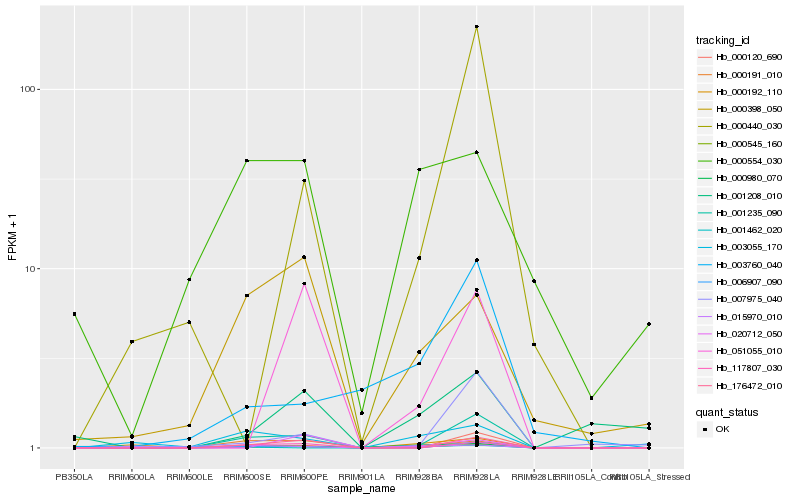
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000545_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 2 |
Hb_001235_090 |
0.1392508163 |
- |
- |
PREDICTED: uncharacterized protein LOC103449952 [Malus domestica] |
| 3 |
Hb_176472_010 |
0.2587723387 |
- |
- |
BnaC01g33060D [Brassica napus] |
| 4 |
Hb_000191_010 |
0.2683417027 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_117807_030 |
0.2779870033 |
- |
- |
Protein MCM10 [Gossypium arboreum] |
| 6 |
Hb_000120_690 |
0.2977594426 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 7 |
Hb_000398_050 |
0.3166894254 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 8 |
Hb_015970_010 |
0.3239862955 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 9 |
Hb_003055_170 |
0.331237932 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 25 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000980_070 |
0.333739858 |
- |
- |
PREDICTED: uncharacterized protein LOC103440883 [Malus domestica] |
| 11 |
Hb_051055_010 |
0.3346697833 |
- |
- |
- |
| 12 |
Hb_003760_040 |
0.3459365466 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 13 |
Hb_001208_010 |
0.3480579435 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF010-like [Jatropha curcas] |
| 14 |
Hb_000192_110 |
0.3529222236 |
- |
- |
PREDICTED: uncharacterized protein LOC105646856 [Jatropha curcas] |
| 15 |
Hb_000554_030 |
0.3533578094 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007975_040 |
0.3548210091 |
- |
- |
PREDICTED: uncharacterized protein LOC104610918 [Nelumbo nucifera] |
| 17 |
Hb_006907_090 |
0.3620388826 |
- |
- |
- |
| 18 |
Hb_020712_050 |
0.3664171885 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 19 |
Hb_000440_030 |
0.3666108597 |
- |
- |
50 kDa protein [Hevea brasiliensis] |
| 20 |
Hb_001462_020 |
0.3701253161 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |