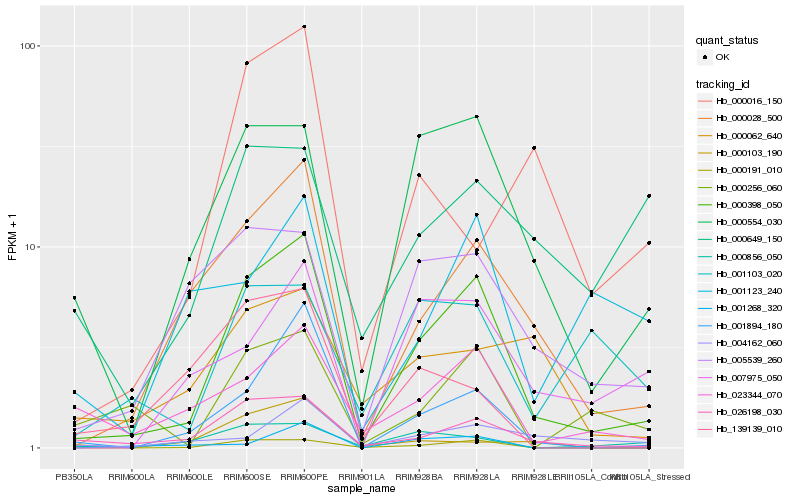
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000398_050 |
0.0 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 2 |
Hb_000028_500 |
0.1912715175 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_000191_010 |
0.1995755277 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_026198_030 |
0.2009814343 |
- |
- |
PREDICTED: ABC transporter G family member 25 [Jatropha curcas] |
| 5 |
Hb_001894_180 |
0.2210961577 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 6 |
Hb_000554_030 |
0.2272657652 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_023344_070 |
0.2460440667 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000256_060 |
0.2582833055 |
- |
- |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 9 |
Hb_005539_260 |
0.2647872893 |
- |
- |
PREDICTED: potassium transporter 6 [Jatropha curcas] |
| 10 |
Hb_007975_050 |
0.2733535651 |
- |
- |
PREDICTED: uncharacterized protein LOC105649339 [Jatropha curcas] |
| 11 |
Hb_000856_050 |
0.275913074 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 12 |
Hb_001268_320 |
0.2880068945 |
- |
- |
PREDICTED: uncharacterized protein LOC104887816 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_004162_060 |
0.2891710865 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_001103_020 |
0.2991943848 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 15 |
Hb_139139_010 |
0.3047557447 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000062_640 |
0.3051743286 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 17 |
Hb_000103_190 |
0.3093733958 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 18 |
Hb_001123_240 |
0.3106686597 |
- |
- |
JAZ10 [Hevea brasiliensis] |
| 19 |
Hb_000016_150 |
0.3128222765 |
- |
- |
PREDICTED: uncharacterized protein LOC105137629 [Populus euphratica] |
| 20 |
Hb_000649_150 |
0.3138745565 |
- |
- |
PREDICTED: uncharacterized protein LOC105634115 [Jatropha curcas] |