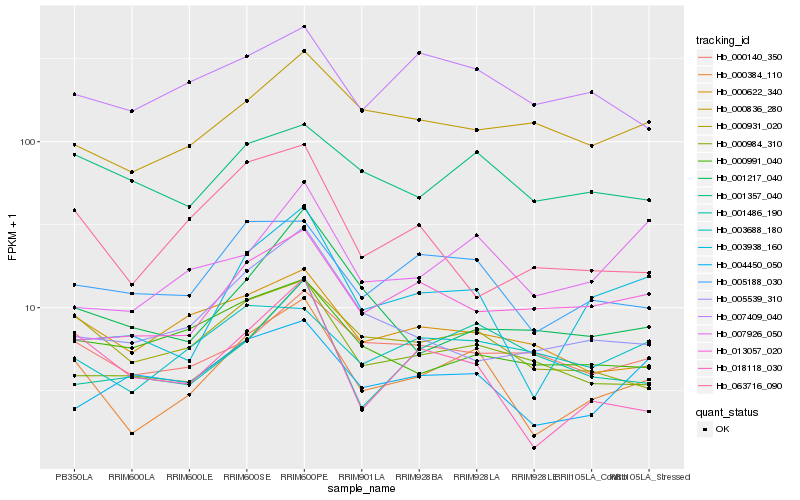
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000384_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003938_160 |
0.155834188 |
- |
- |
PREDICTED: transcription factor bHLH155 [Jatropha curcas] |
| 3 |
Hb_005188_030 |
0.1584879024 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_000931_020 |
0.159060576 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 5 |
Hb_018118_030 |
0.1633319448 |
- |
- |
PREDICTED: putative multidrug resistance protein, partial [Musa acuminata subsp. malaccensis] |
| 6 |
Hb_000984_310 |
0.1768052129 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 7 |
Hb_000622_340 |
0.1820624541 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase MGP4 [Jatropha curcas] |
| 8 |
Hb_007926_050 |
0.1881379701 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 9 |
Hb_001357_040 |
0.1891422373 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 10 |
Hb_000140_350 |
0.1908671383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004450_050 |
0.1919846978 |
- |
- |
PREDICTED: histone chaperone ASF1B [Populus euphratica] |
| 12 |
Hb_000836_280 |
0.1924578823 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005539_310 |
0.1924796481 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 14 |
Hb_003688_180 |
0.1925567992 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 15 |
Hb_013057_020 |
0.1937422046 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 16 |
Hb_063716_090 |
0.1954586605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007409_040 |
0.1969239437 |
- |
- |
actin family protein [Populus trichocarpa] |
| 18 |
Hb_000991_040 |
0.197108466 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 19 |
Hb_001217_040 |
0.1976791709 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 20 |
Hb_001486_190 |
0.1983267686 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |