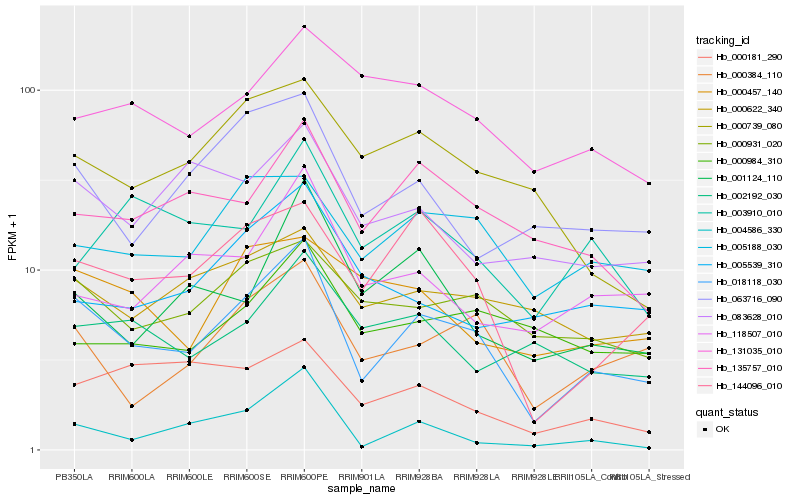
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018118_030 |
0.0 |
- |
- |
PREDICTED: putative multidrug resistance protein, partial [Musa acuminata subsp. malaccensis] |
| 2 |
Hb_000384_110 |
0.1633319448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_144096_010 |
0.1730256623 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 4 |
Hb_000931_020 |
0.1782279188 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005188_030 |
0.1871590428 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_063716_090 |
0.1877154142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_135757_010 |
0.1906136027 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 8 |
Hb_000622_340 |
0.1947600863 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase MGP4 [Jatropha curcas] |
| 9 |
Hb_001124_110 |
0.1978905624 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_083628_010 |
0.1986300677 |
- |
- |
PREDICTED: probable serine protease EDA2 [Jatropha curcas] |
| 11 |
Hb_118507_010 |
0.2022643183 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 12 |
Hb_000739_080 |
0.2024494706 |
- |
- |
PREDICTED: 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase [Jatropha curcas] |
| 13 |
Hb_004586_330 |
0.2036688206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000457_140 |
0.2054033285 |
- |
- |
PREDICTED: transmembrane protein 136-like [Jatropha curcas] |
| 15 |
Hb_002192_030 |
0.2073822336 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_131035_010 |
0.2081058978 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000984_310 |
0.2090703387 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 18 |
Hb_000181_290 |
0.2098848967 |
- |
- |
hypothetical protein POPTR_0001s41000g [Populus trichocarpa] |
| 19 |
Hb_005539_310 |
0.2115773211 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 20 |
Hb_003910_010 |
0.2125390441 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |