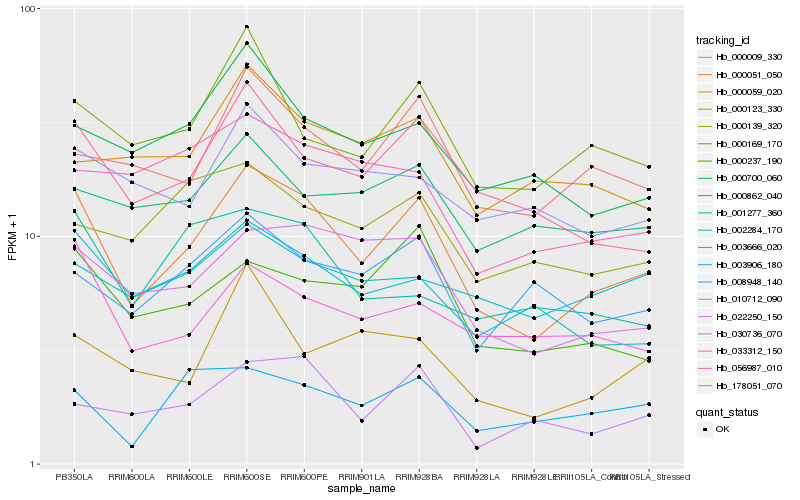
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000051_050 |
0.0 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 2 |
Hb_178051_070 |
0.1221241429 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 3 |
Hb_030736_070 |
0.1273849317 |
- |
- |
PREDICTED: general transcription factor IIH subunit 3 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000009_330 |
0.1345129445 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 5 |
Hb_000169_170 |
0.1349509448 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 6 |
Hb_000237_190 |
0.1367254744 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003666_020 |
0.141541957 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_033312_150 |
0.1421989708 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 9 |
Hb_000700_060 |
0.1441546526 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 10 |
Hb_002284_170 |
0.1477606706 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 11 |
Hb_003906_180 |
0.1483630983 |
- |
- |
unknown [Zea mays] |
| 12 |
Hb_008948_140 |
0.1485472097 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 13 |
Hb_022250_150 |
0.1486940959 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 14 |
Hb_000862_040 |
0.153654351 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 15 |
Hb_010712_090 |
0.1543471679 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 16 |
Hb_000139_320 |
0.1555456092 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 17 |
Hb_056987_010 |
0.1562959125 |
- |
- |
DNA-damage-repair/toleration protein DRT111, chloroplastic [Glycine soja] |
| 18 |
Hb_001277_360 |
0.1563943201 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 19 |
Hb_000123_330 |
0.1568666381 |
- |
- |
hypothetical protein M569_14136, partial [Genlisea aurea] |
| 20 |
Hb_000059_020 |
0.1570197353 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |