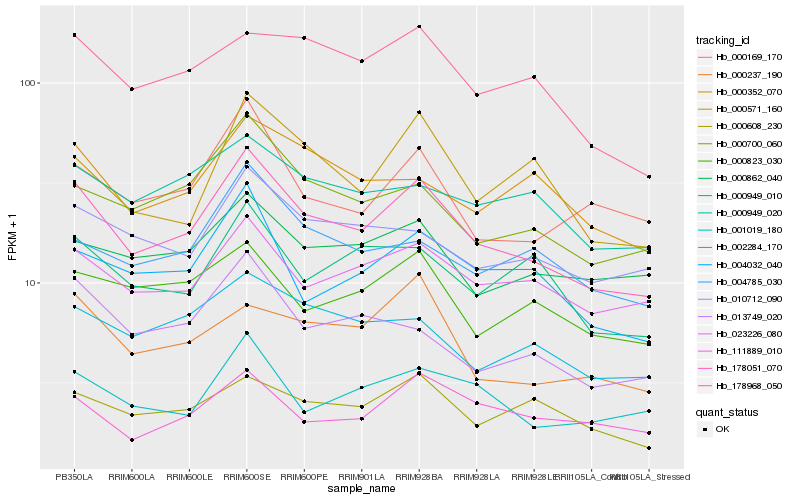
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_178051_070 |
0.0 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 2 |
Hb_000700_060 |
0.0991284582 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 3 |
Hb_023226_080 |
0.1008054563 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004032_040 |
0.1041002081 |
- |
- |
unknown [Medicago truncatula] |
| 5 |
Hb_000169_170 |
0.1072055432 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 6 |
Hb_002284_170 |
0.1080637981 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 7 |
Hb_010712_090 |
0.1085962631 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 8 |
Hb_178968_050 |
0.1088778111 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 9 |
Hb_001019_180 |
0.1093657743 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 10 |
Hb_013749_020 |
0.1100949014 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 11 |
Hb_000949_010 |
0.1109769411 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 12 |
Hb_000571_160 |
0.111582057 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 13 |
Hb_000608_230 |
0.1116375766 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 14 |
Hb_004785_030 |
0.1121983907 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 15 |
Hb_000823_030 |
0.1128117702 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 16 |
Hb_000352_070 |
0.1141169166 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 17 |
Hb_000949_020 |
0.1141489258 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 18 |
Hb_111889_010 |
0.1141641093 |
- |
- |
PREDICTED: DNA ligase 4 [Jatropha curcas] |
| 19 |
Hb_000237_190 |
0.1148184201 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000862_040 |
0.1149085641 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |