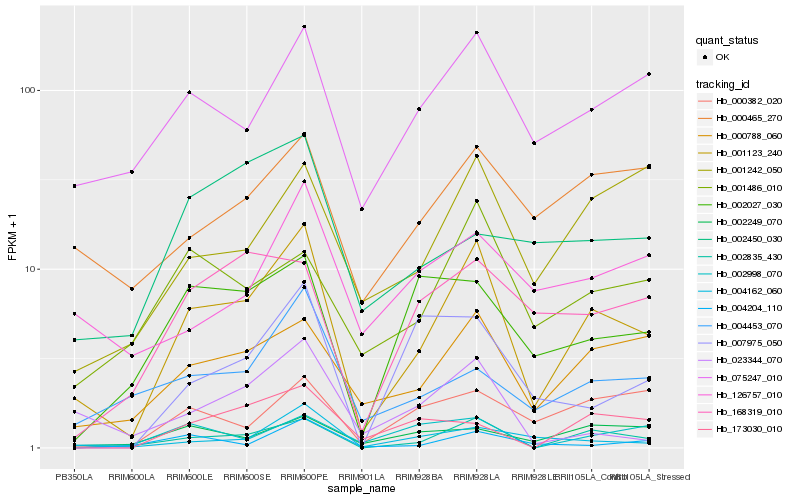
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_240 |
0.0 |
- |
- |
JAZ10 [Hevea brasiliensis] |
| 2 |
Hb_002835_430 |
0.1682310961 |
- |
- |
Alliin lyase precursor, putative [Ricinus communis] |
| 3 |
Hb_000788_060 |
0.2052569598 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |
| 4 |
Hb_075247_010 |
0.2180662486 |
- |
- |
PREDICTED: actin-depolymerizing factor 2-like [Nelumbo nucifera] |
| 5 |
Hb_004162_060 |
0.2323123616 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_004453_070 |
0.2413591297 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_023344_070 |
0.2418029323 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 8 |
Hb_001486_010 |
0.2439099259 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 9 |
Hb_000465_270 |
0.2472684906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001242_050 |
0.2495716489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004204_110 |
0.2560204656 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |
| 12 |
Hb_002027_030 |
0.2571041734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000382_020 |
0.2582042676 |
- |
- |
PREDICTED: uncharacterized protein LOC105633610 [Jatropha curcas] |
| 14 |
Hb_002249_070 |
0.2607620292 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 15 |
Hb_126757_010 |
0.261506884 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 16 |
Hb_168319_010 |
0.261932183 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002998_070 |
0.2645076646 |
- |
- |
hypothetical protein CARUB_v10019139mg, partial [Capsella rubella] |
| 18 |
Hb_007975_050 |
0.2679430607 |
- |
- |
PREDICTED: uncharacterized protein LOC105649339 [Jatropha curcas] |
| 19 |
Hb_173030_010 |
0.2718979521 |
- |
- |
hypothetical protein RCOM_1321060 [Ricinus communis] |
| 20 |
Hb_002450_030 |
0.2722292069 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |