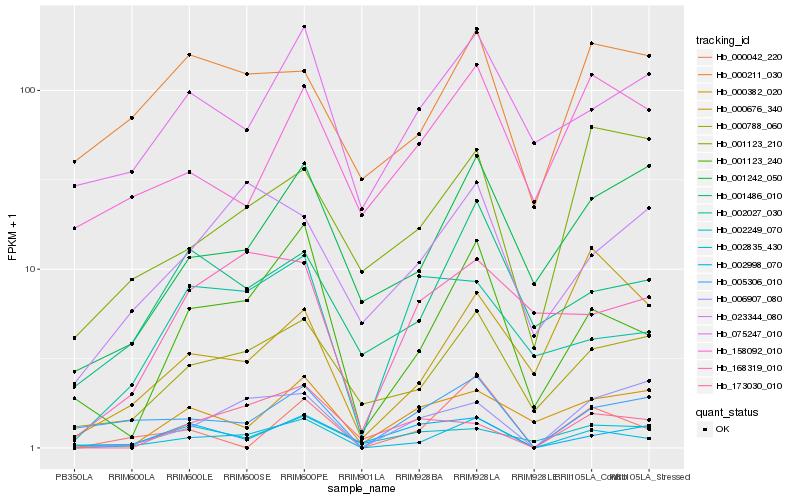
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002835_430 |
0.0 |
- |
- |
Alliin lyase precursor, putative [Ricinus communis] |
| 2 |
Hb_001123_240 |
0.1682310961 |
- |
- |
JAZ10 [Hevea brasiliensis] |
| 3 |
Hb_000788_060 |
0.2331288916 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |
| 4 |
Hb_005306_010 |
0.2523477335 |
- |
- |
- |
| 5 |
Hb_000042_220 |
0.2589493116 |
- |
- |
PREDICTED: uncharacterized protein LOC105632809 [Jatropha curcas] |
| 6 |
Hb_006907_080 |
0.2590314862 |
- |
- |
PREDICTED: RPM1-interacting protein 4-like [Jatropha curcas] |
| 7 |
Hb_001242_050 |
0.2653147215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001486_010 |
0.277784386 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 9 |
Hb_173030_010 |
0.2786312843 |
- |
- |
hypothetical protein RCOM_1321060 [Ricinus communis] |
| 10 |
Hb_075247_010 |
0.2794299313 |
- |
- |
PREDICTED: actin-depolymerizing factor 2-like [Nelumbo nucifera] |
| 11 |
Hb_000382_020 |
0.2866699486 |
- |
- |
PREDICTED: uncharacterized protein LOC105633610 [Jatropha curcas] |
| 12 |
Hb_023344_080 |
0.2882070217 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002998_070 |
0.2886564265 |
- |
- |
hypothetical protein CARUB_v10019139mg, partial [Capsella rubella] |
| 14 |
Hb_158092_010 |
0.2935861665 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 15 |
Hb_168319_010 |
0.2937519626 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002249_070 |
0.2937919572 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 17 |
Hb_001123_210 |
0.2946023153 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |
| 18 |
Hb_002027_030 |
0.2972997636 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000211_030 |
0.2974882955 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 20 |
Hb_000676_340 |
0.2979265448 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |