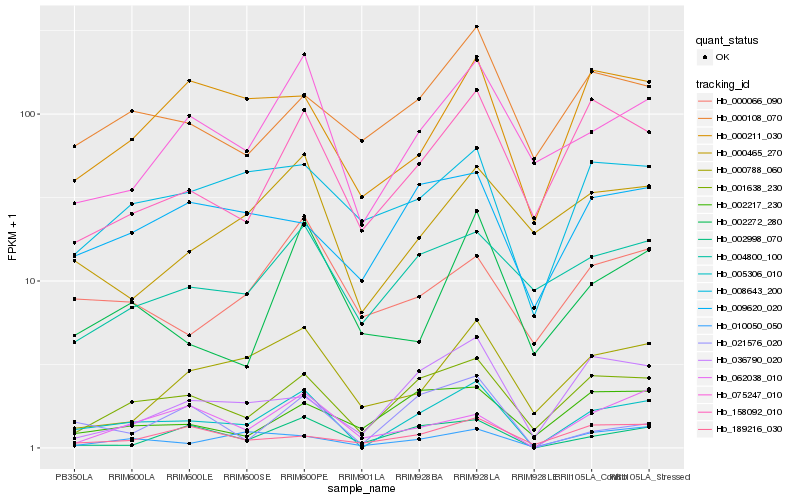
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005306_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001638_230 |
0.18222181 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 3 |
Hb_075247_010 |
0.2143922303 |
- |
- |
PREDICTED: actin-depolymerizing factor 2-like [Nelumbo nucifera] |
| 4 |
Hb_002998_070 |
0.225120336 |
- |
- |
hypothetical protein CARUB_v10019139mg, partial [Capsella rubella] |
| 5 |
Hb_010050_050 |
0.228726525 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog isoform X2 [Jatropha curcas] |
| 6 |
Hb_158092_010 |
0.2288617004 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 7 |
Hb_000211_030 |
0.2335365815 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 8 |
Hb_008643_200 |
0.2340633518 |
transcription factor |
TF Family: BES1 |
PREDICTED: BES1/BZR1 homolog protein 4 [Vitis vinifera] |
| 9 |
Hb_021576_020 |
0.2366965479 |
- |
- |
PREDICTED: uncharacterized protein LOC104426132 [Eucalyptus grandis] |
| 10 |
Hb_036790_020 |
0.2383334965 |
- |
- |
PREDICTED: polyadenylate-binding protein 3 [Jatropha curcas] |
| 11 |
Hb_062038_010 |
0.2396676547 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC104882266 isoform X2 [Vitis vinifera] |
| 12 |
Hb_000788_060 |
0.2407534649 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |
| 13 |
Hb_002272_280 |
0.2408013533 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 14 |
Hb_009620_020 |
0.2416696946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_189216_030 |
0.2433422794 |
- |
- |
Pleiotropic drug resistance protein, putative [Ricinus communis] |
| 16 |
Hb_004800_100 |
0.2452552889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000465_270 |
0.2456062244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000066_090 |
0.2459113562 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 19 |
Hb_002217_230 |
0.2461388754 |
- |
- |
PREDICTED: uncharacterized protein LOC105643583 [Jatropha curcas] |
| 20 |
Hb_000108_070 |
0.2464788084 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |