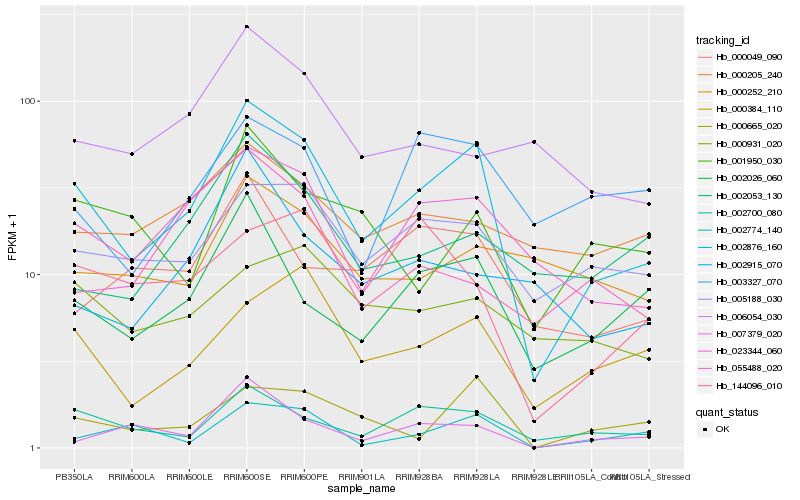
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_160 |
0.0 |
- |
- |
sucrose transporter 1 [Hevea brasiliensis] |
| 2 |
Hb_002700_080 |
0.1799019774 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |
| 3 |
Hb_002026_060 |
0.1884093576 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 4 |
Hb_005188_030 |
0.1952936317 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_023344_060 |
0.1996822274 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA11 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000384_110 |
0.2022007426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000665_020 |
0.2070517931 |
- |
- |
- |
| 8 |
Hb_000049_090 |
0.217715314 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002053_130 |
0.2213095731 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 10 |
Hb_000931_020 |
0.2285764217 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002774_140 |
0.2287181728 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001950_030 |
0.2288096171 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 13 |
Hb_000205_240 |
0.2294047347 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_003327_070 |
0.2295694022 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 15 |
Hb_144096_010 |
0.2305230905 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 16 |
Hb_000252_210 |
0.2314286634 |
- |
- |
PREDICTED: uncharacterized protein LOC105644655 [Jatropha curcas] |
| 17 |
Hb_006054_030 |
0.2315750889 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 18 |
Hb_007379_020 |
0.2321356054 |
- |
- |
hypothetical protein JCGZ_25958 [Jatropha curcas] |
| 19 |
Hb_055488_020 |
0.2323935566 |
- |
- |
PREDICTED: uncharacterized protein LOC105641270 [Jatropha curcas] |
| 20 |
Hb_002915_070 |
0.2333754559 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |