| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
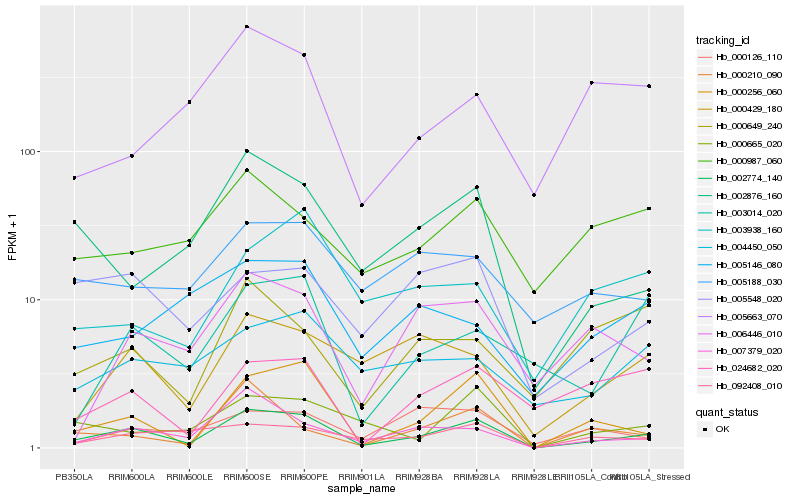
Hb_002774_140 |
0.0 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000256_060 |
0.1885631584 |
- |
- |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 3 |
Hb_007379_020 |
0.2139331142 |
- |
- |
hypothetical protein JCGZ_25958 [Jatropha curcas] |
| 4 |
Hb_002876_160 |
0.2287181728 |
- |
- |
sucrose transporter 1 [Hevea brasiliensis] |
| 5 |
Hb_092408_010 |
0.2362789268 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ and MATH domain-containing protein 3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000429_180 |
0.2380924539 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000665_020 |
0.2400986155 |
- |
- |
- |
| 8 |
Hb_005548_020 |
0.2420553839 |
- |
- |
hypothetical protein JCGZ_05605 [Jatropha curcas] |
| 9 |
Hb_024682_020 |
0.249440962 |
- |
- |
hypothetical protein POPTR_0001s12710g [Populus trichocarpa] |
| 10 |
Hb_000210_090 |
0.2502467274 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 11 |
Hb_003014_020 |
0.2506292089 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 12 |
Hb_006446_010 |
0.2511922201 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_004450_050 |
0.2540142665 |
- |
- |
PREDICTED: histone chaperone ASF1B [Populus euphratica] |
| 14 |
Hb_005188_030 |
0.258365184 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_000987_060 |
0.261710128 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 16 |
Hb_000649_240 |
0.2621046813 |
- |
- |
PREDICTED: probable peroxygenase 4 [Jatropha curcas] |
| 17 |
Hb_003938_160 |
0.2631843006 |
- |
- |
PREDICTED: transcription factor bHLH155 [Jatropha curcas] |
| 18 |
Hb_000126_110 |
0.2652243954 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X3 [Populus euphratica] |
| 19 |
Hb_005663_070 |
0.2662613856 |
- |
- |
dehydrin protein [Manihot esculenta] |
| 20 |
Hb_005146_080 |
0.2671087523 |
- |
- |
PREDICTED: uncharacterized protein LOC105636704 isoform X2 [Jatropha curcas] |