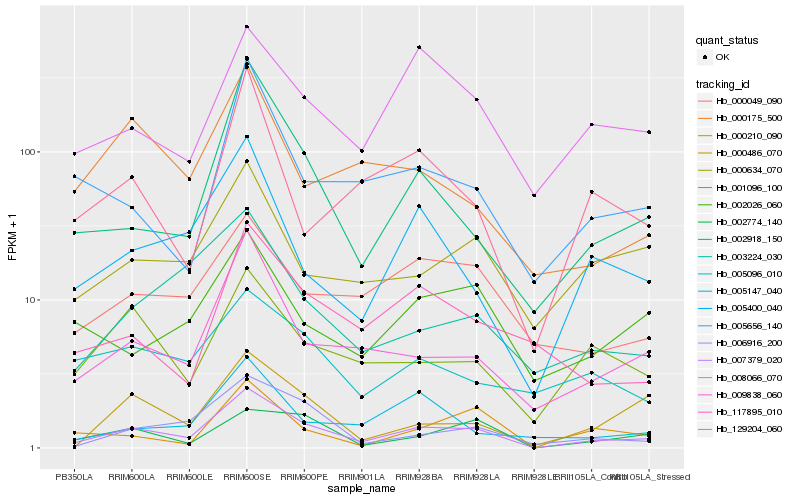
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007379_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_25958 [Jatropha curcas] |
| 2 |
Hb_009838_060 |
0.1832482766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002918_150 |
0.1848365318 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 1 [Hevea brasiliensis] |
| 4 |
Hb_005400_040 |
0.2001558728 |
- |
- |
PREDICTED: uncharacterized protein At4g04775-like [Jatropha curcas] |
| 5 |
Hb_006916_200 |
0.2047323809 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 6 |
Hb_001096_100 |
0.2048599086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_008066_070 |
0.2109819294 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 8 |
Hb_000049_090 |
0.2123792603 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002774_140 |
0.2139331142 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_117895_010 |
0.2147470554 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 11 |
Hb_000634_070 |
0.2169329353 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 12 |
Hb_000486_070 |
0.217211162 |
- |
- |
hypothetical protein JCGZ_25069 [Jatropha curcas] |
| 13 |
Hb_005656_140 |
0.2173296496 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 14 |
Hb_003224_030 |
0.2186675293 |
- |
- |
hypothetical protein POPTR_0077s002402g, partial [Populus trichocarpa] |
| 15 |
Hb_129204_060 |
0.2209120213 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 16 |
Hb_005147_040 |
0.2236130797 |
- |
- |
PREDICTED: uncharacterized protein LOC105629674 [Jatropha curcas] |
| 17 |
Hb_002026_060 |
0.2242204045 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 18 |
Hb_000210_090 |
0.2247788106 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 19 |
Hb_005096_010 |
0.2276238405 |
- |
- |
PREDICTED: uncharacterized protein LOC105641403 [Jatropha curcas] |
| 20 |
Hb_000175_500 |
0.2280749437 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |