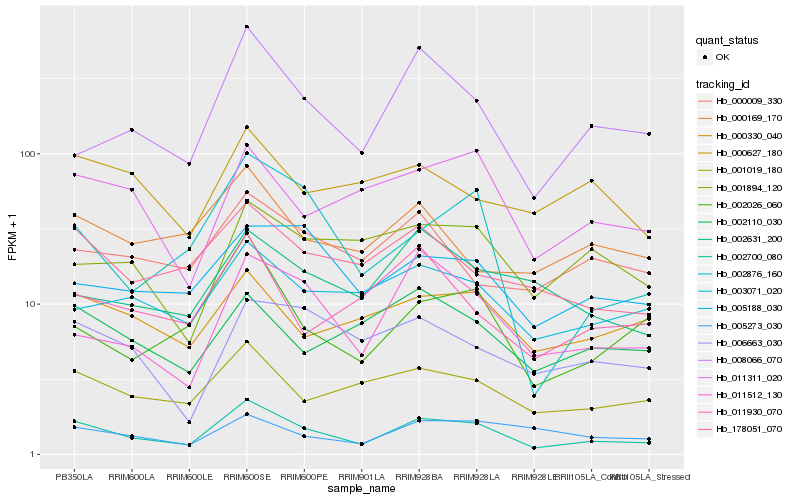
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002700_080 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |
| 2 |
Hb_001019_180 |
0.1575537628 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 3 |
Hb_011512_130 |
0.16425339 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 4 |
Hb_008066_070 |
0.1661045405 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 5 |
Hb_178051_070 |
0.1666721453 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 6 |
Hb_002026_060 |
0.1667342625 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 7 |
Hb_011311_020 |
0.167669749 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003071_020 |
0.1723672023 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 9 |
Hb_000330_040 |
0.1738755353 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 10 |
Hb_002110_030 |
0.1797284094 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 11 |
Hb_002876_160 |
0.1799019774 |
- |
- |
sucrose transporter 1 [Hevea brasiliensis] |
| 12 |
Hb_005188_030 |
0.180477327 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_011930_070 |
0.1811193157 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 14 |
Hb_006663_030 |
0.1812914584 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 15 |
Hb_005273_030 |
0.1847446877 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 16 |
Hb_000169_170 |
0.1867837773 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 17 |
Hb_002631_200 |
0.1869417995 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001894_120 |
0.1880258906 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 19 |
Hb_000009_330 |
0.1886789084 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_000627_180 |
0.1905530775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |