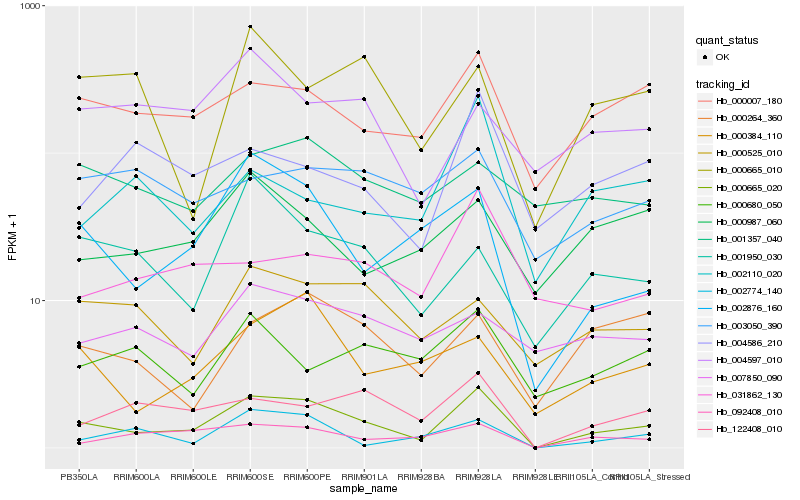
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000665_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_002876_160 |
0.2070517931 |
- |
- |
sucrose transporter 1 [Hevea brasiliensis] |
| 3 |
Hb_000007_180 |
0.2159506599 |
- |
- |
hypothetical protein PRUPE_ppb024585mg, partial [Prunus persica] |
| 4 |
Hb_122408_010 |
0.2162079511 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_092408_010 |
0.2207975803 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ and MATH domain-containing protein 3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000384_110 |
0.228800926 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002774_140 |
0.2400986155 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001950_030 |
0.2409903179 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 9 |
Hb_000264_360 |
0.2455627814 |
- |
- |
hypothetical protein JCGZ_21443 [Jatropha curcas] |
| 10 |
Hb_000525_010 |
0.2460669707 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 11 |
Hb_004586_210 |
0.2483347018 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_031862_130 |
0.2493778619 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000987_060 |
0.2512709857 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 14 |
Hb_004597_010 |
0.2543667266 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor MYC2 [Jatropha curcas] |
| 15 |
Hb_000680_050 |
0.2553114956 |
- |
- |
hypothetical protein JCGZ_24767 [Jatropha curcas] |
| 16 |
Hb_000665_010 |
0.2554427888 |
- |
- |
hypothetical protein 20 [Hevea brasiliensis] |
| 17 |
Hb_002110_020 |
0.2582144801 |
- |
- |
PREDICTED: transcription factor bHLH145 [Jatropha curcas] |
| 18 |
Hb_003050_390 |
0.2585666164 |
- |
- |
PREDICTED: uncharacterized protein LOC104442543 [Eucalyptus grandis] |
| 19 |
Hb_007850_090 |
0.2590245915 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001357_040 |
0.2605923208 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |