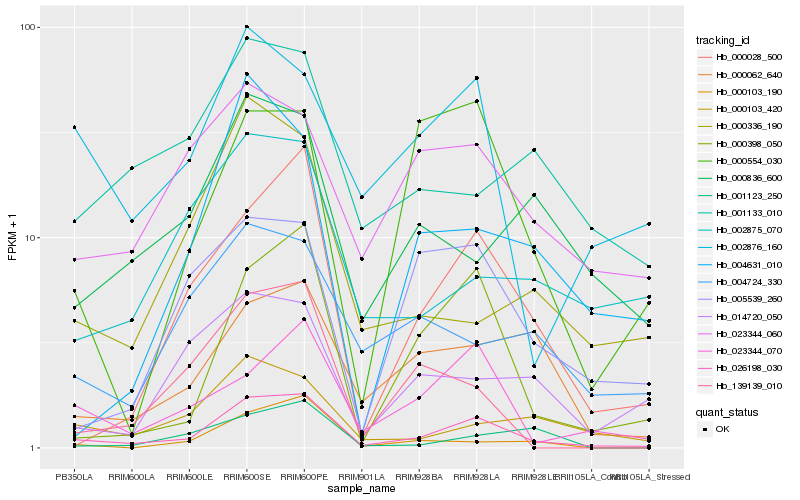
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026198_030 |
0.0 |
- |
- |
PREDICTED: ABC transporter G family member 25 [Jatropha curcas] |
| 2 |
Hb_000398_050 |
0.2009814343 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 3 |
Hb_000028_500 |
0.2021830058 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_023344_070 |
0.2148023252 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000062_640 |
0.2262211808 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |
| 6 |
Hb_000103_420 |
0.233498142 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Populus euphratica] |
| 7 |
Hb_002876_160 |
0.23795995 |
- |
- |
sucrose transporter 1 [Hevea brasiliensis] |
| 8 |
Hb_000103_190 |
0.2431341068 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 9 |
Hb_023344_060 |
0.2465629528 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA11 isoform X2 [Jatropha curcas] |
| 10 |
Hb_139139_010 |
0.2484880033 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005539_260 |
0.2501553157 |
- |
- |
PREDICTED: potassium transporter 6 [Jatropha curcas] |
| 12 |
Hb_004724_330 |
0.2504485093 |
- |
- |
Capsanthin/capsorubin synthase, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_014720_050 |
0.2506150162 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 14 |
Hb_002875_070 |
0.2513953561 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 15 |
Hb_000836_600 |
0.2534368124 |
- |
- |
PREDICTED: delta(8)-fatty-acid desaturase 2 [Jatropha curcas] |
| 16 |
Hb_000336_190 |
0.255273131 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 17 |
Hb_004631_010 |
0.2553258357 |
- |
- |
Early nodulin 20 precursor, putative [Ricinus communis] |
| 18 |
Hb_001133_010 |
0.2560277196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000554_030 |
0.2577749452 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001123_250 |
0.2582724529 |
- |
- |
hypothetical protein POPTR_0001s13220g [Populus trichocarpa] |