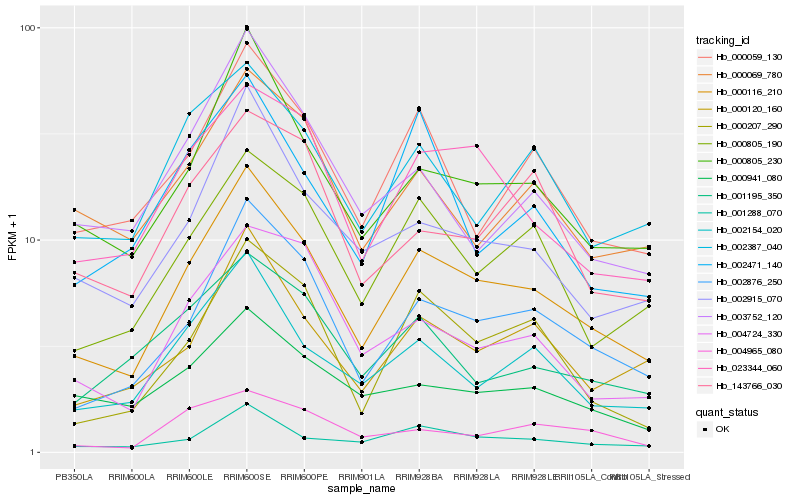
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628567 [Jatropha curcas] |
| 2 |
Hb_002876_250 |
0.085169584 |
- |
- |
PREDICTED: receptor-like protein kinase 2 [Jatropha curcas] |
| 3 |
Hb_000805_230 |
0.1030404449 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 4 |
Hb_001288_070 |
0.1122560515 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 5 |
Hb_000120_160 |
0.1131943686 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_002154_020 |
0.1142476071 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_023344_060 |
0.116137968 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA11 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000805_190 |
0.1165867325 |
transcription factor |
TF Family: GRAS |
PREDICTED: uncharacterized protein LOC103323607 [Prunus mume] |
| 9 |
Hb_002915_070 |
0.1181977738 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 10 |
Hb_000059_130 |
0.1203263608 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 11 |
Hb_000207_290 |
0.1260631848 |
- |
- |
hypothetical protein JCGZ_03920 [Jatropha curcas] |
| 12 |
Hb_002387_040 |
0.1317610296 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 13 |
Hb_000069_780 |
0.1354408583 |
- |
- |
atpob1, putative [Ricinus communis] |
| 14 |
Hb_000941_080 |
0.1361694631 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 7-like isoform X3 [Populus euphratica] |
| 15 |
Hb_004724_330 |
0.1399314626 |
- |
- |
Capsanthin/capsorubin synthase, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_003752_120 |
0.1434022832 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001195_350 |
0.14481247 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 18 |
Hb_004965_080 |
0.1469645053 |
- |
- |
hypothetical protein F383_09654 [Gossypium arboreum] |
| 19 |
Hb_143766_030 |
0.1478148467 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_002471_140 |
0.1485164098 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |