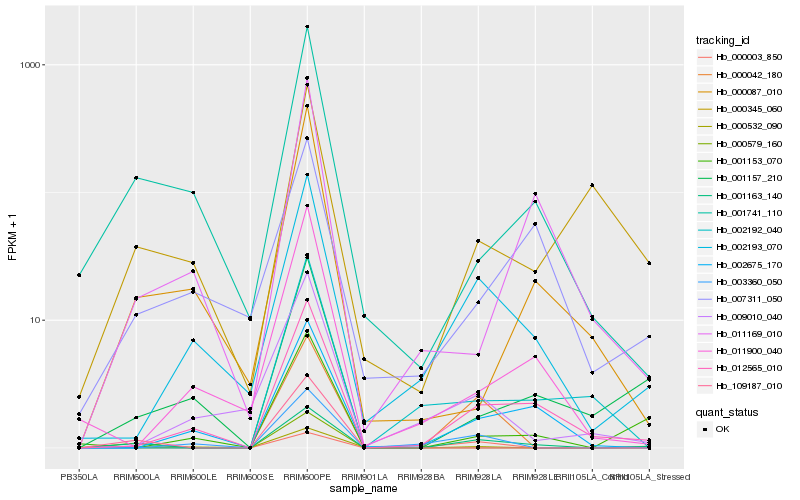
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001163_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_012565_010 |
0.231134344 |
- |
- |
PREDICTED: putative glutamine amidotransferase PB2B2.05 [Jatropha curcas] |
| 3 |
Hb_002193_070 |
0.2412556082 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000003_850 |
0.2494508866 |
- |
- |
PREDICTED: uncharacterized protein LOC105633597 [Jatropha curcas] |
| 5 |
Hb_000042_180 |
0.260081428 |
- |
- |
- |
| 6 |
Hb_001157_210 |
0.2685762823 |
- |
- |
hypothetical protein CISIN_1g038260mg [Citrus sinensis] |
| 7 |
Hb_002675_170 |
0.2726812144 |
- |
- |
hypothetical protein JCGZ_06235 [Jatropha curcas] |
| 8 |
Hb_001741_110 |
0.2726976086 |
rubber biosynthesis |
Gene Name: REF7 |
REF/SRPP-like protein [Glycine soja] |
| 9 |
Hb_001153_070 |
0.2857835181 |
- |
- |
- |
| 10 |
Hb_000532_090 |
0.2858263378 |
- |
- |
ATPase [delta proteobacterium MLMS-1] |
| 11 |
Hb_007311_050 |
0.2978786913 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000087_010 |
0.2997878938 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 13 |
Hb_011900_040 |
0.3001808173 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 14 |
Hb_011169_010 |
0.3013375277 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 15 |
Hb_002192_040 |
0.3031994924 |
- |
- |
- |
| 16 |
Hb_003360_050 |
0.3046783688 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 17 |
Hb_000345_060 |
0.3048264633 |
- |
- |
PREDICTED: REF/SRPP-like protein At3g05500 [Jatropha curcas] |
| 18 |
Hb_000579_160 |
0.305951155 |
- |
- |
PREDICTED: uncharacterized protein LOC104216549 isoform X1 [Nicotiana sylvestris] |
| 19 |
Hb_109187_010 |
0.3061226831 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Cucumis sativus] |
| 20 |
Hb_009010_040 |
0.3109800474 |
- |
- |
Peptide transporter, putative [Ricinus communis] |