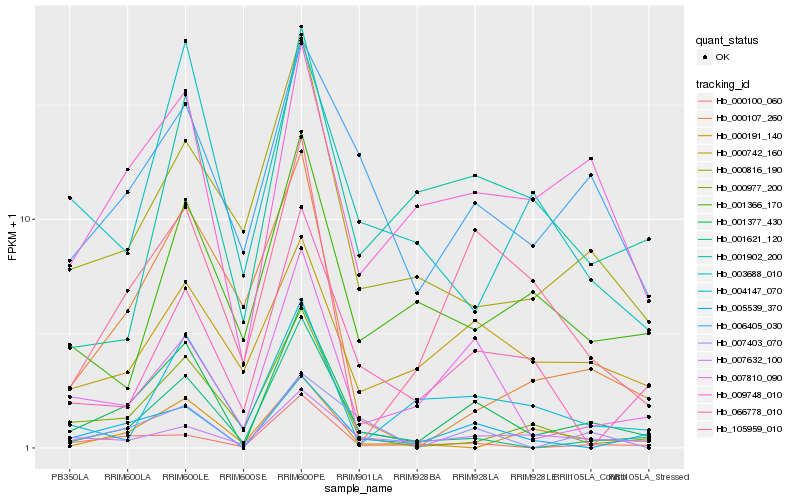
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_430 |
0.0 |
- |
- |
PREDICTED: laccase-6 [Jatropha curcas] |
| 2 |
Hb_005539_370 |
0.2027132884 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 3 |
Hb_001621_120 |
0.2063650343 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 4 |
Hb_007810_090 |
0.2083732041 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |
| 5 |
Hb_009748_010 |
0.2163997527 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 6 |
Hb_105959_010 |
0.2279751861 |
- |
- |
asparagine synthetase, putative [Ricinus communis] |
| 7 |
Hb_000100_060 |
0.2301978289 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Populus euphratica] |
| 8 |
Hb_006405_030 |
0.2359224297 |
- |
- |
hypothetical protein VITISV_043238 [Vitis vinifera] |
| 9 |
Hb_007403_070 |
0.239204362 |
- |
- |
hypothetical protein JCGZ_23672 [Jatropha curcas] |
| 10 |
Hb_000977_200 |
0.2404018438 |
- |
- |
PREDICTED: acyl-[acyl-carrier-protein] desaturase, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_007632_100 |
0.240659206 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 12 |
Hb_000816_190 |
0.2412302695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000742_160 |
0.2524603623 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g20940 [Jatropha curcas] |
| 14 |
Hb_000107_260 |
0.2551064101 |
- |
- |
- |
| 15 |
Hb_001902_200 |
0.2573104345 |
- |
- |
PREDICTED: tubulin beta-1 chain [Eucalyptus grandis] |
| 16 |
Hb_066778_010 |
0.257579974 |
- |
- |
- |
| 17 |
Hb_004147_070 |
0.262338051 |
- |
- |
PREDICTED: probable rhamnogalacturonate lyase B [Jatropha curcas] |
| 18 |
Hb_001366_170 |
0.2655204711 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 19 |
Hb_003688_010 |
0.2678406551 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3-like [Jatropha curcas] |
| 20 |
Hb_000191_140 |
0.2682080881 |
- |
- |
PREDICTED: growth-regulating factor 1-like isoform X2 [Jatropha curcas] |