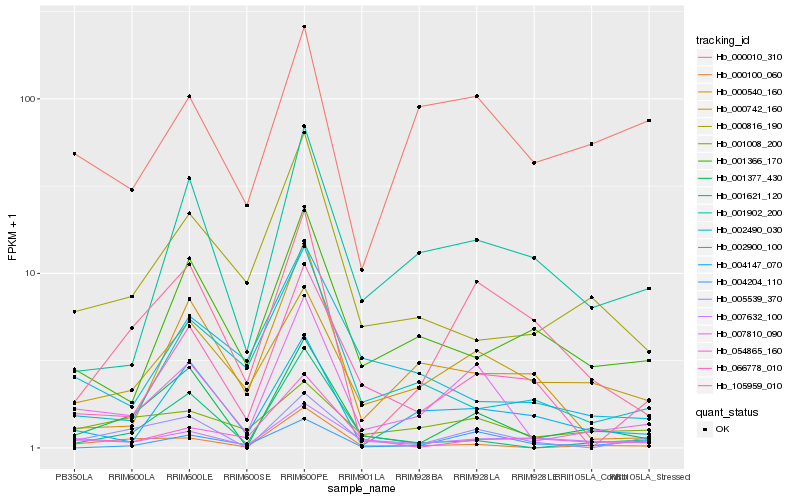
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007810_090 |
0.0 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |
| 2 |
Hb_001377_430 |
0.2083732041 |
- |
- |
PREDICTED: laccase-6 [Jatropha curcas] |
| 3 |
Hb_000100_060 |
0.2194103899 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Populus euphratica] |
| 4 |
Hb_005539_370 |
0.224990574 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 5 |
Hb_000742_160 |
0.2258424758 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g20940 [Jatropha curcas] |
| 6 |
Hb_007632_100 |
0.2258579547 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 7 |
Hb_066778_010 |
0.2290372764 |
- |
- |
- |
| 8 |
Hb_105959_010 |
0.2342329745 |
- |
- |
asparagine synthetase, putative [Ricinus communis] |
| 9 |
Hb_001902_200 |
0.2346988186 |
- |
- |
PREDICTED: tubulin beta-1 chain [Eucalyptus grandis] |
| 10 |
Hb_004147_070 |
0.2360579148 |
- |
- |
PREDICTED: probable rhamnogalacturonate lyase B [Jatropha curcas] |
| 11 |
Hb_000816_190 |
0.2432265371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004204_110 |
0.2434618001 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |
| 13 |
Hb_001008_200 |
0.2461712358 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC103961552 [Pyrus x bretschneideri] |
| 14 |
Hb_001366_170 |
0.251602034 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 15 |
Hb_001621_120 |
0.2559673012 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 16 |
Hb_000010_310 |
0.25687491 |
- |
- |
PREDICTED: probable pectate lyase 18 [Populus euphratica] |
| 17 |
Hb_054865_160 |
0.2580877051 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 18 |
Hb_002490_030 |
0.259777888 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000540_160 |
0.2602798981 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF6 [Jatropha curcas] |
| 20 |
Hb_002900_100 |
0.2619365977 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |