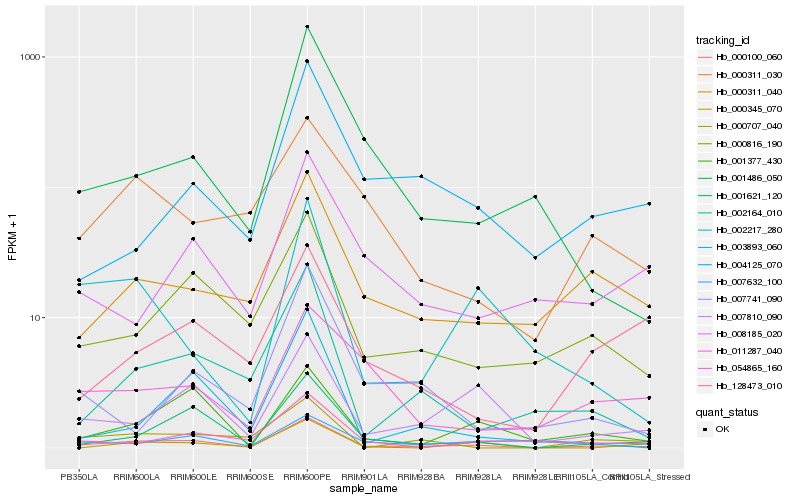
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000100_060 |
0.0 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Populus euphratica] |
| 2 |
Hb_001621_120 |
0.1836718338 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 3 |
Hb_000816_190 |
0.2171143535 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007810_090 |
0.2194103899 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |
| 5 |
Hb_002164_010 |
0.2264668637 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 6 |
Hb_001377_430 |
0.2301978289 |
- |
- |
PREDICTED: laccase-6 [Jatropha curcas] |
| 7 |
Hb_000311_040 |
0.2403574154 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 8 |
Hb_054865_160 |
0.2470695805 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 9 |
Hb_001486_050 |
0.2479270751 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 10 |
Hb_003893_060 |
0.2522230343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000707_040 |
0.2564550762 |
- |
- |
Uncharacterized protein TCM_021829 [Theobroma cacao] |
| 12 |
Hb_008185_020 |
0.2596115291 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 13 |
Hb_002217_280 |
0.2625237291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007741_090 |
0.2637762244 |
- |
- |
Autophagy-related 8f -like protein [Gossypium arboreum] |
| 15 |
Hb_004125_070 |
0.2643566566 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 16 |
Hb_128473_010 |
0.2649355143 |
- |
- |
- |
| 17 |
Hb_000311_030 |
0.2666956411 |
- |
- |
hypothetical protein B456_002G227600 [Gossypium raimondii] |
| 18 |
Hb_000345_070 |
0.2674511187 |
rubber biosynthesis |
Gene Name: SRPP6 |
- |
| 19 |
Hb_007632_100 |
0.2685117588 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 20 |
Hb_011287_040 |
0.2687103115 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |