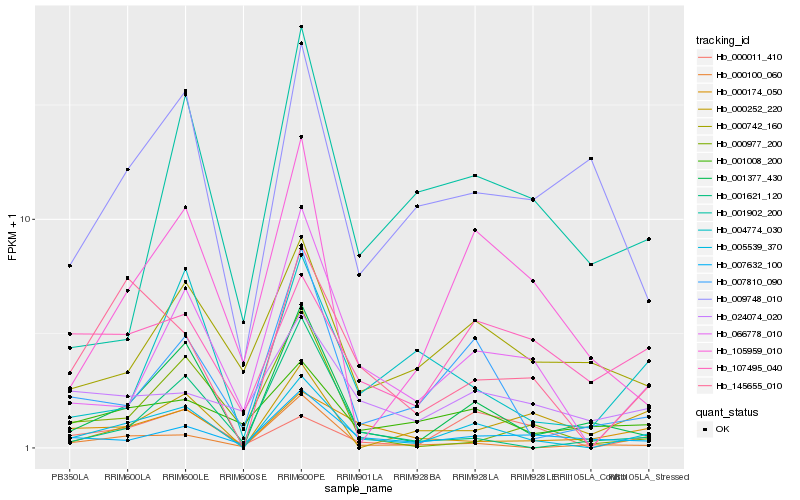
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005539_370 |
0.0 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 2 |
Hb_001377_430 |
0.2027132884 |
- |
- |
PREDICTED: laccase-6 [Jatropha curcas] |
| 3 |
Hb_145655_010 |
0.2076719011 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 4 |
Hb_066778_010 |
0.2151267657 |
- |
- |
- |
| 5 |
Hb_007810_090 |
0.224990574 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |
| 6 |
Hb_007632_100 |
0.2578155219 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 7 |
Hb_004774_030 |
0.2665948779 |
- |
- |
PREDICTED: uncharacterized protein LOC105648177 [Jatropha curcas] |
| 8 |
Hb_000174_050 |
0.2688322061 |
- |
- |
PREDICTED: gamma-tubulin complex component 2 [Populus euphratica] |
| 9 |
Hb_000100_060 |
0.270717383 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Populus euphratica] |
| 10 |
Hb_000011_410 |
0.2709876719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_105959_010 |
0.2719680416 |
- |
- |
asparagine synthetase, putative [Ricinus communis] |
| 12 |
Hb_001008_200 |
0.2756434007 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC103961552 [Pyrus x bretschneideri] |
| 13 |
Hb_000742_160 |
0.2808989218 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g20940 [Jatropha curcas] |
| 14 |
Hb_107495_040 |
0.2847345214 |
- |
- |
PREDICTED: 12-oxophytodienoate reductase 2-like [Citrus sinensis] |
| 15 |
Hb_009748_010 |
0.2856867799 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 16 |
Hb_001621_120 |
0.2868481041 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 17 |
Hb_001902_200 |
0.2878938985 |
- |
- |
PREDICTED: tubulin beta-1 chain [Eucalyptus grandis] |
| 18 |
Hb_024074_020 |
0.2884562702 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 19 |
Hb_000977_200 |
0.2918760253 |
- |
- |
PREDICTED: acyl-[acyl-carrier-protein] desaturase, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_000252_220 |
0.2920651929 |
- |
- |
PREDICTED: copper amine oxidase 1-like isoform X2 [Beta vulgaris subsp. vulgaris] |