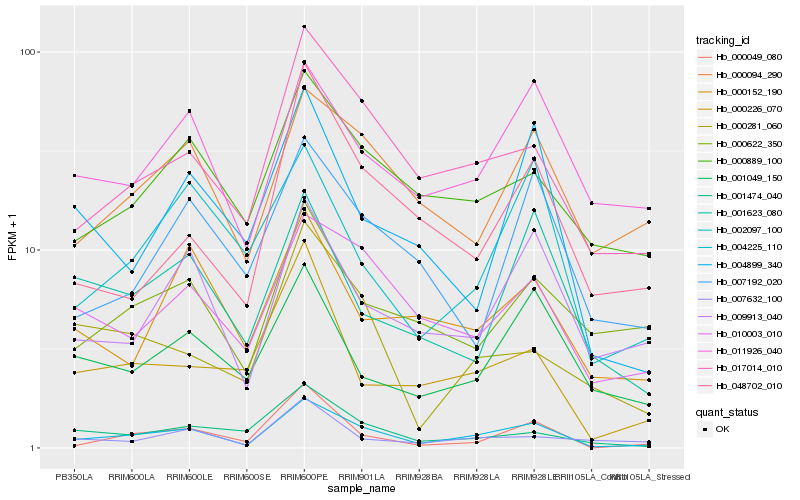
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004225_110 |
0.0 |
- |
- |
PREDICTED: ABC transporter G family member 28 [Jatropha curcas] |
| 2 |
Hb_017014_010 |
0.1602160146 |
- |
- |
PREDICTED: syntaxin-132-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_009913_040 |
0.1798696092 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 4 |
Hb_010003_010 |
0.181424402 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000049_080 |
0.1907445946 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000889_100 |
0.1908424549 |
- |
- |
PREDICTED: syntaxin-132-like isoform X2 [Sesamum indicum] |
| 7 |
Hb_000094_290 |
0.1909708392 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_011926_040 |
0.1915453089 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001049_150 |
0.2020745631 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 10 |
Hb_002097_100 |
0.2028523332 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_048702_010 |
0.2031973237 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_001474_040 |
0.2042081605 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 13 |
Hb_001623_080 |
0.2069200228 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 14 |
Hb_000281_060 |
0.2071432914 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000622_350 |
0.2072792588 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 16 |
Hb_000226_070 |
0.2082961567 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 17 |
Hb_004899_340 |
0.2083707937 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 18 |
Hb_007632_100 |
0.2102578242 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 19 |
Hb_000152_190 |
0.211282036 |
- |
- |
PREDICTED: K(+) efflux antiporter 5 [Jatropha curcas] |
| 20 |
Hb_007192_020 |
0.2125534436 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |