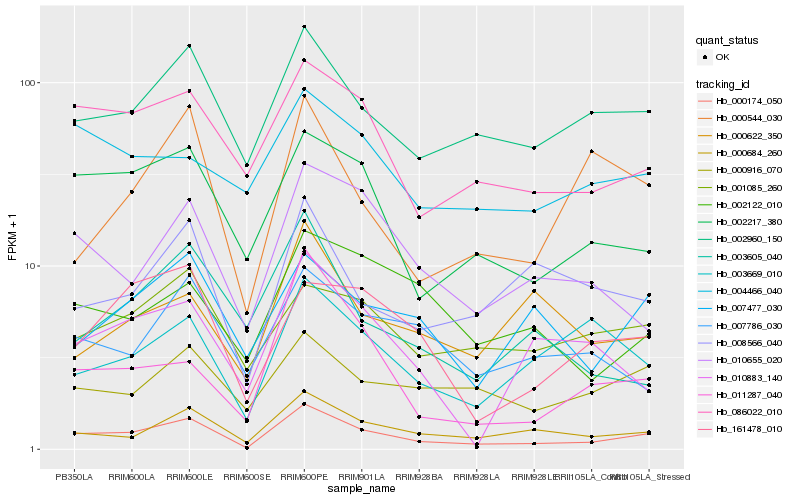
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000174_050 |
0.0 |
- |
- |
PREDICTED: gamma-tubulin complex component 2 [Populus euphratica] |
| 2 |
Hb_000684_260 |
0.1513106857 |
- |
- |
PREDICTED: probable DNA helicase MCM8 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002960_150 |
0.1654732942 |
- |
- |
hypothetical protein CISIN_1g029215mg [Citrus sinensis] |
| 4 |
Hb_002217_380 |
0.171119627 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 5 |
Hb_086022_010 |
0.1718766171 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 6 |
Hb_000916_070 |
0.1898909893 |
- |
- |
PREDICTED: sister chromatid cohesion protein DCC1-like [Jatropha curcas] |
| 7 |
Hb_010655_020 |
0.1905778314 |
- |
- |
hypothetical protein CICLE_v10032410mg [Citrus clementina] |
| 8 |
Hb_008566_040 |
0.1942659228 |
- |
- |
PREDICTED: uncharacterized protein LOC105647590 isoform X1 [Jatropha curcas] |
| 9 |
Hb_161478_010 |
0.1960780419 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_003669_010 |
0.1979434221 |
- |
- |
PREDICTED: psbP domain-containing protein 5, chloroplastic [Populus euphratica] |
| 11 |
Hb_001085_260 |
0.1988344846 |
- |
- |
PREDICTED: uncharacterized protein LOC105633184 [Jatropha curcas] |
| 12 |
Hb_007477_030 |
0.1995589891 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 13 |
Hb_000544_030 |
0.200758772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011287_040 |
0.2044176673 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_007786_030 |
0.205452843 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Jatropha curcas] |
| 16 |
Hb_002122_010 |
0.2105055753 |
- |
- |
- |
| 17 |
Hb_010883_140 |
0.2107222623 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000622_350 |
0.2133249881 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 19 |
Hb_004466_040 |
0.2135094664 |
- |
- |
PREDICTED: protein RER1B-like [Gossypium raimondii] |
| 20 |
Hb_003605_040 |
0.2146343364 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 isoform X1 [Jatropha curcas] |