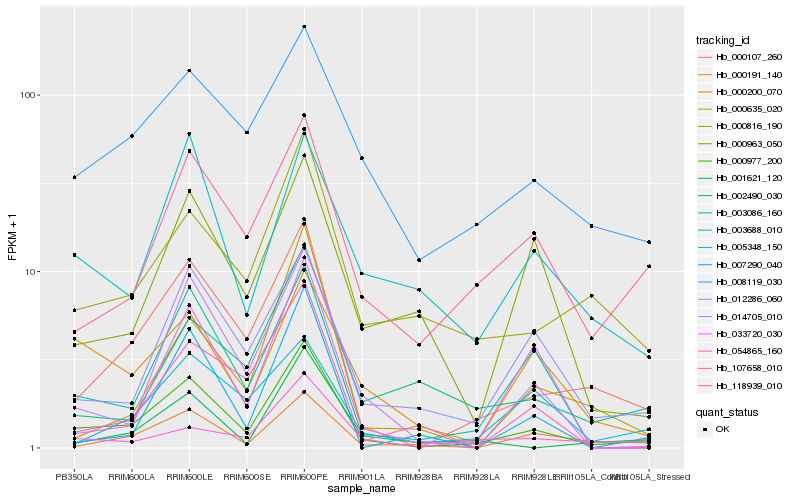
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000977_200 |
0.0 |
- |
- |
PREDICTED: acyl-[acyl-carrier-protein] desaturase, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000107_260 |
0.1480358395 |
- |
- |
- |
| 3 |
Hb_003086_160 |
0.1836001125 |
- |
- |
hypothetical protein POPTR_0006s21590g [Populus trichocarpa] |
| 4 |
Hb_012286_060 |
0.1906078387 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_014705_010 |
0.197604329 |
- |
- |
hypothetical protein JCGZ_21106 [Jatropha curcas] |
| 6 |
Hb_000191_140 |
0.2000601073 |
- |
- |
PREDICTED: growth-regulating factor 1-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000816_190 |
0.2017383233 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_118939_010 |
0.2053539272 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_008119_030 |
0.2056149176 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 10 |
Hb_054865_160 |
0.2102565034 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 11 |
Hb_000200_070 |
0.2146380109 |
- |
- |
PREDICTED: CDP-diacylglycerol--serine O-phosphatidyltransferase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001621_120 |
0.2164931985 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 13 |
Hb_003688_010 |
0.2169747692 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3-like [Jatropha curcas] |
| 14 |
Hb_033720_030 |
0.2174741899 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_000635_020 |
0.2198791954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_107658_010 |
0.2228493355 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_007290_040 |
0.2237930125 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1-like [Jatropha curcas] |
| 18 |
Hb_000963_050 |
0.2242297774 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |
| 19 |
Hb_002490_030 |
0.2248918636 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005348_150 |
0.2268700641 |
- |
- |
PREDICTED: actin-100-like [Jatropha curcas] |