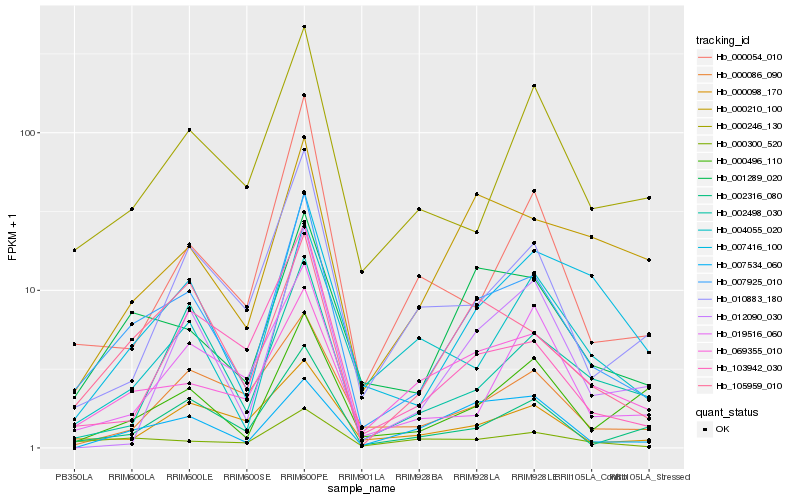
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007925_010 |
0.0 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 1 [Jatropha curcas] |
| 2 |
Hb_001289_020 |
0.1505203613 |
- |
- |
PREDICTED: uncharacterized protein LOC105635827 [Jatropha curcas] |
| 3 |
Hb_105959_010 |
0.1542488987 |
- |
- |
asparagine synthetase, putative [Ricinus communis] |
| 4 |
Hb_002316_080 |
0.1734545715 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 5 |
Hb_069355_010 |
0.1914704484 |
- |
- |
NBS-LRR resistance protein RGH2 [Manihot esculenta] |
| 6 |
Hb_007416_100 |
0.1937544883 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 5 [Jatropha curcas] |
| 7 |
Hb_000246_130 |
0.1949420699 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 8 |
Hb_010883_180 |
0.1963661563 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 9 |
Hb_000098_170 |
0.1983309655 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 10 |
Hb_000300_520 |
0.2029612811 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 11 |
Hb_000496_110 |
0.2036239281 |
- |
- |
PREDICTED: uncharacterized protein LOC105650214 [Jatropha curcas] |
| 12 |
Hb_103942_030 |
0.2041582883 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_000086_090 |
0.2062783832 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 14 |
Hb_002498_030 |
0.2126985959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_012090_030 |
0.2127189293 |
- |
- |
PREDICTED: autophagy-related protein 8C isoform X2 [Phoenix dactylifera] |
| 16 |
Hb_000210_100 |
0.2150011183 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 17 |
Hb_000054_010 |
0.2154133277 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004055_020 |
0.2158181702 |
- |
- |
PREDICTED: uncharacterized protein LOC105631433 [Jatropha curcas] |
| 19 |
Hb_007534_060 |
0.2165088248 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_019516_060 |
0.2170985672 |
- |
- |
PREDICTED: uncharacterized protein LOC105634259 [Jatropha curcas] |