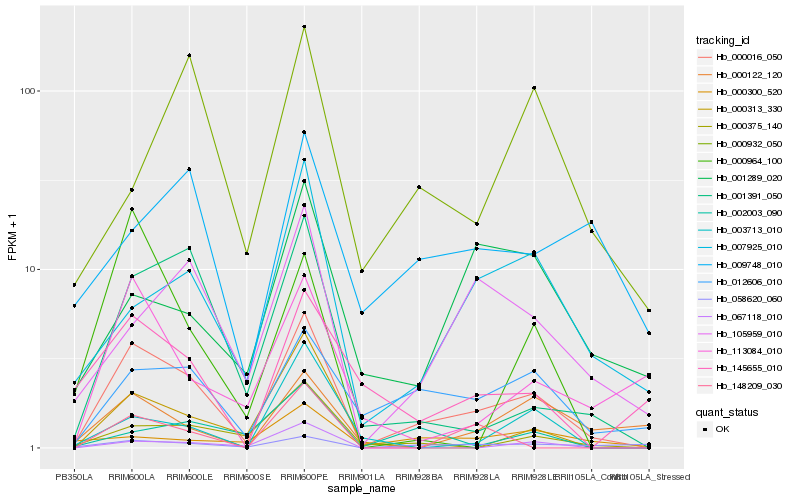
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000016_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648535 [Jatropha curcas] |
| 2 |
Hb_012606_010 |
0.2524414645 |
transcription factor |
TF Family: IWS1 |
PREDICTED: probable mediator of RNA polymerase II transcription subunit 26c isoform X2 [Jatropha curcas] |
| 3 |
Hb_001391_050 |
0.2655072334 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |
| 4 |
Hb_007925_010 |
0.2677546416 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 1 [Jatropha curcas] |
| 5 |
Hb_105959_010 |
0.2697557392 |
- |
- |
asparagine synthetase, putative [Ricinus communis] |
| 6 |
Hb_000375_140 |
0.275270665 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 7 |
Hb_058620_060 |
0.2815088485 |
- |
- |
uncharacterized protein LOC105646159 precursor [Jatropha curcas] |
| 8 |
Hb_067118_010 |
0.2951130815 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 9 |
Hb_113084_010 |
0.2953995424 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 10 |
Hb_145655_010 |
0.2964902638 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 11 |
Hb_000313_330 |
0.2974961566 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase [Jatropha curcas] |
| 12 |
Hb_002003_090 |
0.304862467 |
- |
- |
PREDICTED: TPR repeat-containing protein ZIP4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000964_100 |
0.3094234652 |
- |
- |
PREDICTED: uncharacterized protein LOC105628699 [Jatropha curcas] |
| 14 |
Hb_003713_010 |
0.309601269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000122_120 |
0.3118997661 |
- |
- |
PREDICTED: arogenate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_009748_010 |
0.3133084384 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 17 |
Hb_001289_020 |
0.3137551176 |
- |
- |
PREDICTED: uncharacterized protein LOC105635827 [Jatropha curcas] |
| 18 |
Hb_000300_520 |
0.3141382546 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 19 |
Hb_148209_030 |
0.3146029515 |
- |
- |
hypothetical protein VITISV_017120 [Vitis vinifera] |
| 20 |
Hb_000932_050 |
0.3152162639 |
- |
- |
annexin [Manihot esculenta] |