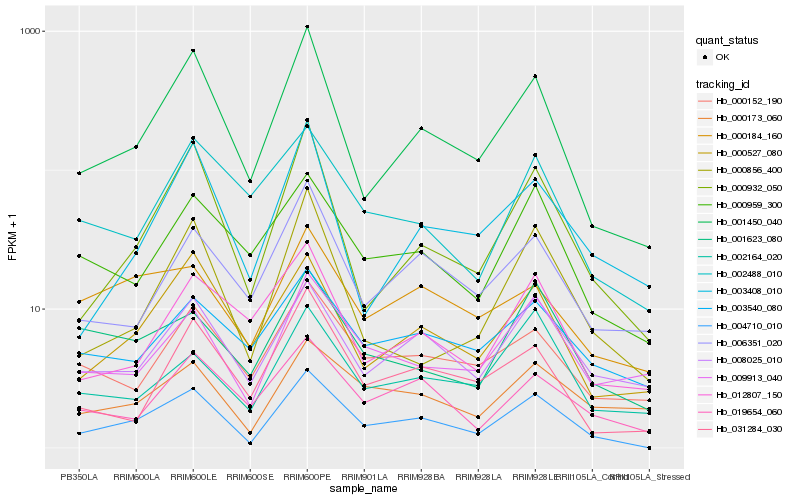
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004710_010 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_D010351, partial [Eucalyptus grandis] |
| 2 |
Hb_001450_040 |
0.1389990107 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 3 |
Hb_000932_050 |
0.1643374959 |
- |
- |
annexin [Manihot esculenta] |
| 4 |
Hb_008025_010 |
0.1690825086 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Musa acuminata subsp. malaccensis] |
| 5 |
Hb_000527_080 |
0.1697884631 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 6 |
Hb_009913_040 |
0.1813133051 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 7 |
Hb_019654_060 |
0.1846482864 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000959_300 |
0.1862927625 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 9 |
Hb_000173_060 |
0.1879904051 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 10 |
Hb_031284_030 |
0.1883136399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000152_190 |
0.1892179917 |
- |
- |
PREDICTED: K(+) efflux antiporter 5 [Jatropha curcas] |
| 12 |
Hb_006351_020 |
0.190060684 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 13 |
Hb_000856_400 |
0.1907344158 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 14 |
Hb_001623_080 |
0.1933971212 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 15 |
Hb_002164_020 |
0.1959649471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000184_160 |
0.1980702495 |
- |
- |
PREDICTED: uncharacterized protein LOC105647991 [Jatropha curcas] |
| 17 |
Hb_003540_080 |
0.1986885775 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_003408_010 |
0.1999601712 |
- |
- |
fructokinase [Manihot esculenta] |
| 19 |
Hb_002488_010 |
0.201456153 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 20 |
Hb_012807_150 |
0.2020162188 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |