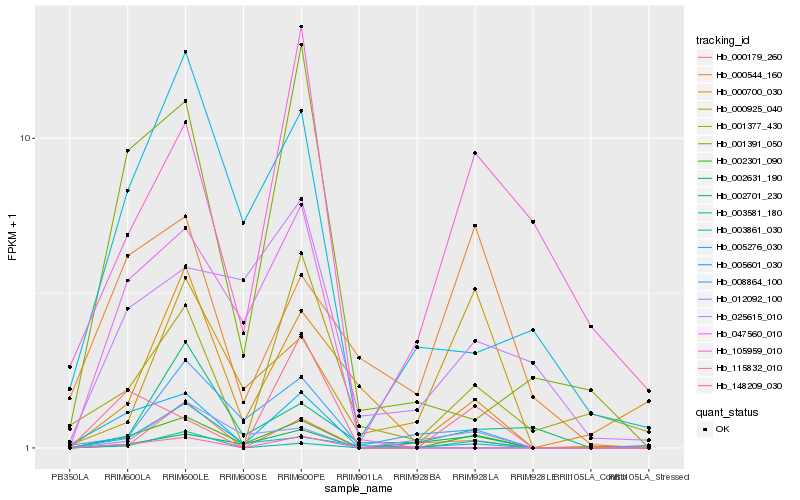
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002301_090 |
0.0 |
- |
- |
PREDICTED: dnaJ protein homolog 1-like [Jatropha curcas] |
| 2 |
Hb_002631_190 |
0.2639744286 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 3 |
Hb_003581_180 |
0.2676998765 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 4 |
Hb_012092_100 |
0.2701723659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008864_100 |
0.2766953605 |
- |
- |
PREDICTED: uncharacterized protein LOC105631832 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005276_030 |
0.292859437 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 7 |
Hb_003861_030 |
0.2971363338 |
- |
- |
hypothetical protein JCGZ_16321 [Jatropha curcas] |
| 8 |
Hb_000925_040 |
0.3032477533 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_047560_010 |
0.3042090586 |
- |
- |
PREDICTED: GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 10 |
Hb_005601_030 |
0.3054299485 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 11 |
Hb_000179_260 |
0.3088027273 |
- |
- |
PREDICTED: UBP1-associated proteins 1C isoform X1 [Jatropha curcas] |
| 12 |
Hb_001391_050 |
0.3145217391 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |
| 13 |
Hb_115832_010 |
0.3190874137 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000700_030 |
0.3301938305 |
- |
- |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 15 |
Hb_025615_010 |
0.3303261238 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 16 |
Hb_001377_430 |
0.3324311522 |
- |
- |
PREDICTED: laccase-6 [Jatropha curcas] |
| 17 |
Hb_105959_010 |
0.3325057056 |
- |
- |
asparagine synthetase, putative [Ricinus communis] |
| 18 |
Hb_148209_030 |
0.3330528308 |
- |
- |
hypothetical protein VITISV_017120 [Vitis vinifera] |
| 19 |
Hb_002701_230 |
0.3330659742 |
- |
- |
PREDICTED: uncharacterized protein LOC105646833 [Jatropha curcas] |
| 20 |
Hb_000544_160 |
0.3346282823 |
- |
- |
PREDICTED: phosphomethylethanolamine N-methyltransferase-like [Nelumbo nucifera] |