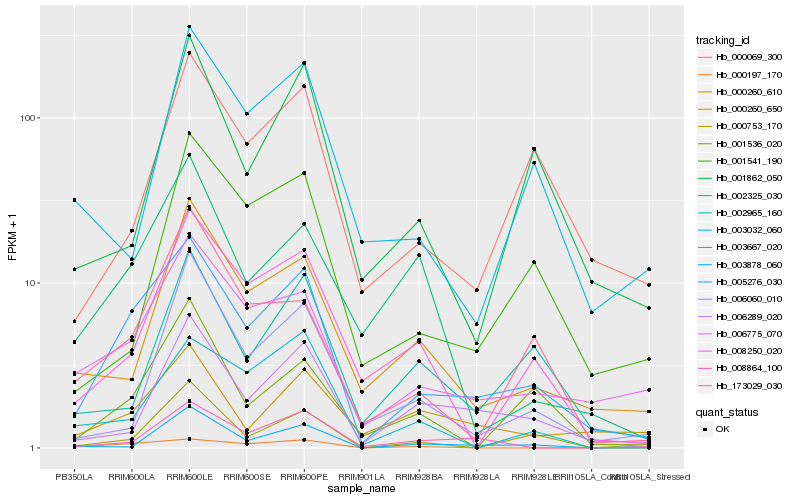
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005276_030 |
0.0 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 2 |
Hb_006775_070 |
0.1744910177 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_008250_020 |
0.2150979691 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_173029_030 |
0.2175139288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000069_300 |
0.2204584328 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000197_170 |
0.2215040258 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_001536_020 |
0.2215696844 |
transcription factor |
TF Family: GRF |
PREDICTED: uncharacterized protein LOC105631006 [Jatropha curcas] |
| 8 |
Hb_006289_020 |
0.2219602676 |
- |
- |
PREDICTED: shugoshin-1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000260_610 |
0.2240755827 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 10 |
Hb_000753_170 |
0.227331989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001541_190 |
0.2316886161 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 12 |
Hb_008864_100 |
0.2324527932 |
- |
- |
PREDICTED: uncharacterized protein LOC105631832 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002965_160 |
0.2338766176 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 14 |
Hb_003032_060 |
0.2351302875 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 15 |
Hb_000260_650 |
0.2370121722 |
- |
- |
PREDICTED: protein PFC0760c [Jatropha curcas] |
| 16 |
Hb_001862_050 |
0.2370395889 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 17 |
Hb_002325_030 |
0.2441411185 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 2 [Jatropha curcas] |
| 18 |
Hb_006060_010 |
0.2441696216 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 19 |
Hb_003878_060 |
0.245099744 |
- |
- |
hypothetical protein B456_007G326900, partial [Gossypium raimondii] |
| 20 |
Hb_003667_020 |
0.246535173 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |