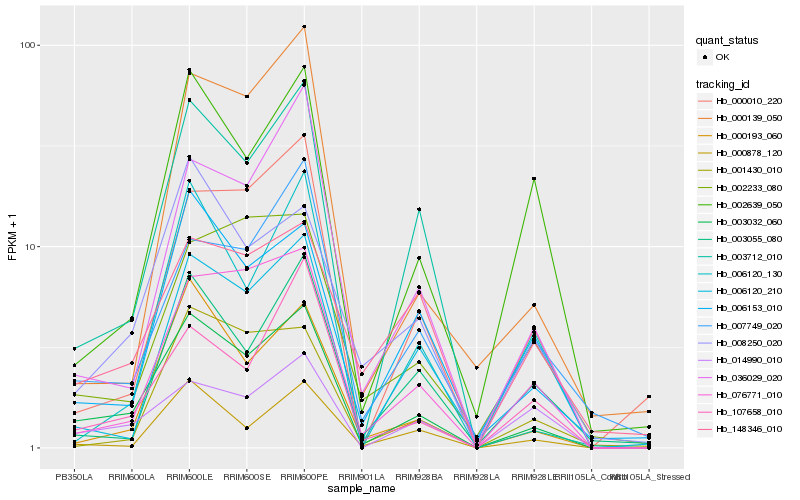
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003032_060 |
0.0 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 2 |
Hb_003712_010 |
0.152984541 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_000193_060 |
0.158520667 |
- |
- |
PREDICTED: F-box protein At4g35930 [Jatropha curcas] |
| 4 |
Hb_006153_010 |
0.1629665729 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 [Populus euphratica] |
| 5 |
Hb_076771_010 |
0.167346285 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 6 |
Hb_008250_020 |
0.1704751205 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000878_120 |
0.1757469564 |
- |
- |
PREDICTED: protein downstream neighbor of Son [Jatropha curcas] |
| 8 |
Hb_036029_020 |
0.1786778094 |
- |
- |
PREDICTED: patellin-3-like [Jatropha curcas] |
| 9 |
Hb_002639_050 |
0.1817710547 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 10 |
Hb_006120_130 |
0.1846083635 |
- |
- |
- |
| 11 |
Hb_000010_220 |
0.1876480973 |
- |
- |
PREDICTED: uncharacterized protein LOC105640277 [Jatropha curcas] |
| 12 |
Hb_007749_020 |
0.1880431639 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 13 |
Hb_107658_010 |
0.1894903262 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_014990_010 |
0.1907678664 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CICLE_v100031791mg, partial [Citrus clementina] |
| 15 |
Hb_001430_010 |
0.191584232 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003055_080 |
0.1927619023 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000139_050 |
0.1955777 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 18 |
Hb_006120_210 |
0.2033677252 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 19 |
Hb_148346_010 |
0.2043676916 |
- |
- |
- |
| 20 |
Hb_002233_080 |
0.2044166469 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |