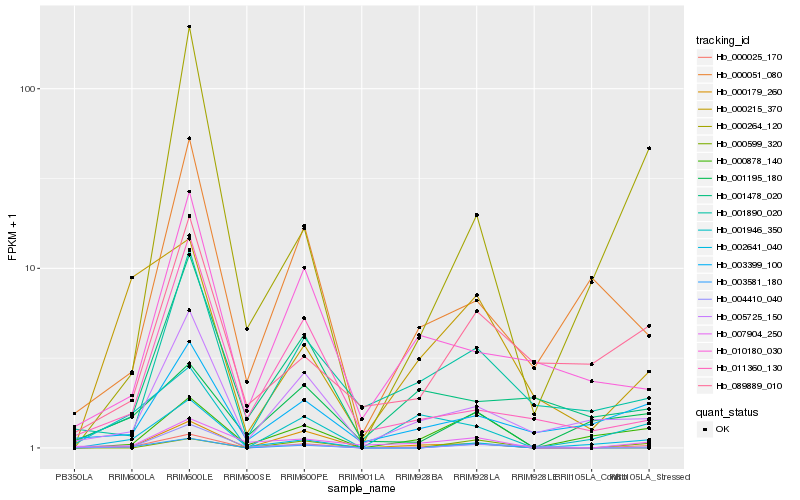
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000878_140 |
0.2531233597 |
- |
- |
PREDICTED: cell division cycle 20.2, cofactor of APC complex-like [Jatropha curcas] |
| 3 |
Hb_003581_180 |
0.2536555111 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 4 |
Hb_005725_150 |
0.2547503052 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001946_350 |
0.2659555399 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 6 |
Hb_007904_250 |
0.2720774707 |
- |
- |
PREDICTED: vignain-like [Jatropha curcas] |
| 7 |
Hb_001890_020 |
0.275276787 |
- |
- |
hypothetical protein CICLE_v10025198mg [Citrus clementina] |
| 8 |
Hb_002641_040 |
0.2763553075 |
- |
- |
class III peroxidase [Populus trichocarpa] |
| 9 |
Hb_000179_260 |
0.2799930426 |
- |
- |
PREDICTED: UBP1-associated proteins 1C isoform X1 [Jatropha curcas] |
| 10 |
Hb_003399_100 |
0.2898132311 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 11 |
Hb_000264_120 |
0.2931274032 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 12 |
Hb_000599_320 |
0.2933074706 |
- |
- |
hypothetical protein JCGZ_17662 [Jatropha curcas] |
| 13 |
Hb_000215_370 |
0.3000453249 |
- |
- |
myoinositol oxygenase, putative [Ricinus communis] |
| 14 |
Hb_001478_020 |
0.3010669912 |
desease resistance |
Gene Name: AAA |
thyroid hormone receptor interactor, putative [Ricinus communis] |
| 15 |
Hb_000051_080 |
0.3029748892 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT3 [Jatropha curcas] |
| 16 |
Hb_010180_030 |
0.3052867848 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 17 |
Hb_089889_010 |
0.3075168127 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_011360_130 |
0.3077811587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004410_040 |
0.3094718137 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_001195_180 |
0.3117352366 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |