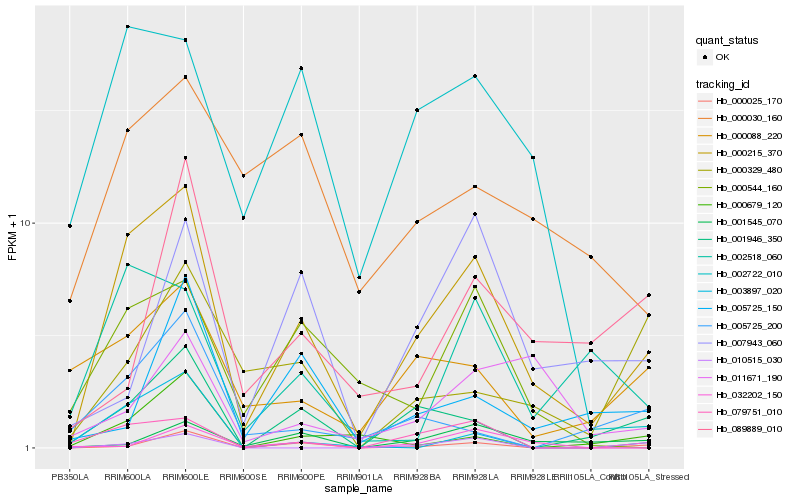
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000215_370 |
0.0 |
- |
- |
myoinositol oxygenase, putative [Ricinus communis] |
| 2 |
Hb_001946_350 |
0.2597080109 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 3 |
Hb_002722_010 |
0.2937053166 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |
| 4 |
Hb_002518_060 |
0.2946180703 |
- |
- |
PREDICTED: uncharacterized protein LOC105631129 [Jatropha curcas] |
| 5 |
Hb_000544_160 |
0.2969398276 |
- |
- |
PREDICTED: phosphomethylethanolamine N-methyltransferase-like [Nelumbo nucifera] |
| 6 |
Hb_079751_010 |
0.2975243734 |
- |
- |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000025_170 |
0.3000453249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000088_220 |
0.3003874019 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_003897_020 |
0.3142740816 |
transcription factor |
TF Family: Orphans |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_000679_120 |
0.3151743484 |
- |
- |
hypothetical protein POPTR_0018s01610g [Populus trichocarpa] |
| 11 |
Hb_000030_160 |
0.3169312355 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 7-like [Jatropha curcas] |
| 12 |
Hb_005725_200 |
0.3188917509 |
transcription factor |
TF Family: GRF |
hypothetical protein JCGZ_15989 [Jatropha curcas] |
| 13 |
Hb_001545_070 |
0.326154596 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_032202_150 |
0.3328416859 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR14 [Jatropha curcas] |
| 15 |
Hb_000329_480 |
0.3367095369 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 4 [Jatropha curcas] |
| 16 |
Hb_010515_030 |
0.3380205108 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 17 |
Hb_007943_060 |
0.3403001933 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 18 |
Hb_089889_010 |
0.3425242229 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 19 |
Hb_005725_150 |
0.3450968977 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 20 |
Hb_011671_190 |
0.3456387664 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |