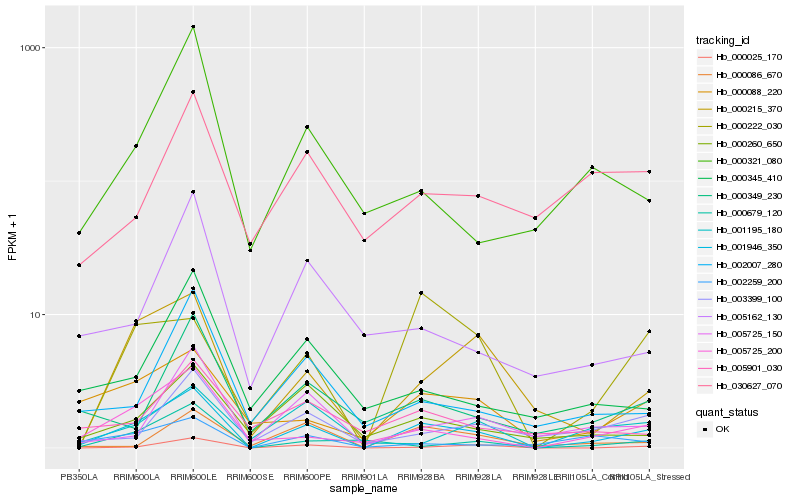
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001946_350 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 2 |
Hb_005725_200 |
0.2464864155 |
transcription factor |
TF Family: GRF |
hypothetical protein JCGZ_15989 [Jatropha curcas] |
| 3 |
Hb_000215_370 |
0.2597080109 |
- |
- |
myoinositol oxygenase, putative [Ricinus communis] |
| 4 |
Hb_000025_170 |
0.2659555399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000088_220 |
0.2694148649 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_005725_150 |
0.2743903118 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003399_100 |
0.2772117861 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 8 |
Hb_000260_650 |
0.2810809601 |
- |
- |
PREDICTED: protein PFC0760c [Jatropha curcas] |
| 9 |
Hb_000086_670 |
0.2877944213 |
- |
- |
PREDICTED: probable DNA helicase MCM9 [Jatropha curcas] |
| 10 |
Hb_000349_230 |
0.2915749986 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 11 |
Hb_002259_200 |
0.292179097 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000222_030 |
0.2927901695 |
- |
- |
- |
| 13 |
Hb_002007_280 |
0.2935493154 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 14 |
Hb_001195_180 |
0.2953150408 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |
| 15 |
Hb_005901_030 |
0.2965072249 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000679_120 |
0.2981722411 |
- |
- |
hypothetical protein POPTR_0018s01610g [Populus trichocarpa] |
| 17 |
Hb_030627_070 |
0.2983441517 |
- |
- |
histone H4 [Zea mays] |
| 18 |
Hb_000345_410 |
0.3014881134 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 19 |
Hb_000321_080 |
0.3039010734 |
- |
- |
histone H4 [Zea mays] |
| 20 |
Hb_005162_130 |
0.3070646174 |
- |
- |
PREDICTED: uncharacterized protein LOC105648433 isoform X2 [Jatropha curcas] |