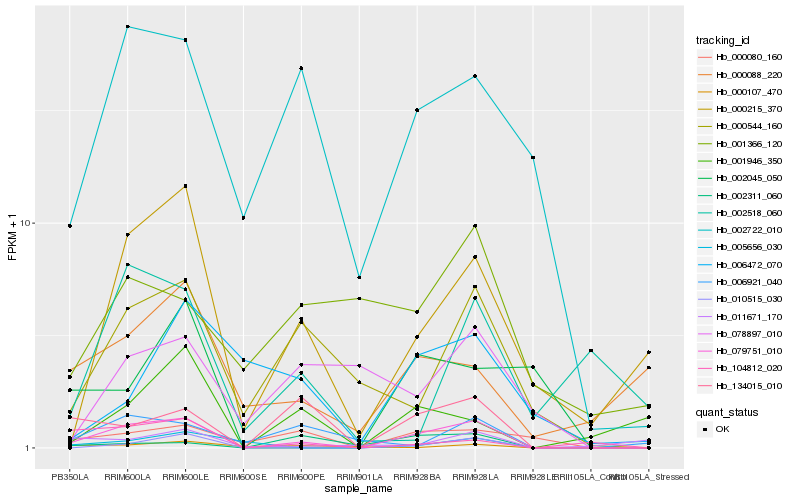
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_079751_010 |
0.0 |
- |
- |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 2 |
Hb_010515_030 |
0.2816138536 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 3 |
Hb_000215_370 |
0.2975243734 |
- |
- |
myoinositol oxygenase, putative [Ricinus communis] |
| 4 |
Hb_002722_010 |
0.3090426315 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |
| 5 |
Hb_134015_010 |
0.3135291982 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 6 |
Hb_000544_160 |
0.3163968022 |
- |
- |
PREDICTED: phosphomethylethanolamine N-methyltransferase-like [Nelumbo nucifera] |
| 7 |
Hb_000080_160 |
0.3324780736 |
- |
- |
Peroxin 13 [Theobroma cacao] |
| 8 |
Hb_005656_030 |
0.3435455187 |
transcription factor |
TF Family: MYB-related |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 9 |
Hb_104812_020 |
0.3517463901 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000107_470 |
0.3538526727 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 11 |
Hb_000088_220 |
0.359035351 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_001946_350 |
0.3635784169 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 13 |
Hb_006921_040 |
0.3638394287 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 14 |
Hb_078897_010 |
0.3766851401 |
- |
- |
hypothetical protein POPTR_0012s044902g, partial [Populus trichocarpa] |
| 15 |
Hb_002311_060 |
0.3770109299 |
- |
- |
PREDICTED: protein trichome birefringence-like 4 [Jatropha curcas] |
| 16 |
Hb_002045_050 |
0.3804476615 |
- |
- |
- |
| 17 |
Hb_002518_060 |
0.3820231669 |
- |
- |
PREDICTED: uncharacterized protein LOC105631129 [Jatropha curcas] |
| 18 |
Hb_006472_070 |
0.3850550817 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 19 |
Hb_011671_170 |
0.3853309045 |
- |
- |
PREDICTED: protein DYAD [Jatropha curcas] |
| 20 |
Hb_001366_120 |
0.3885416969 |
- |
- |
unknown [Zea mays] |