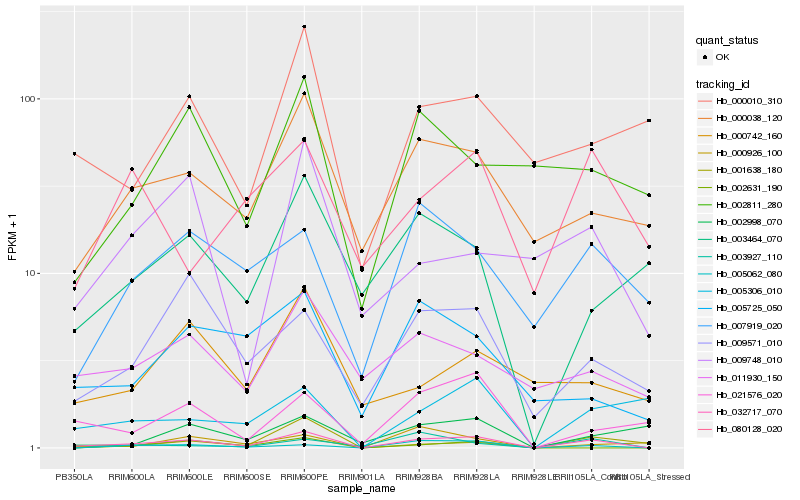
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032717_070 |
0.0 |
- |
- |
PREDICTED: mucin-2-like [Jatropha curcas] |
| 2 |
Hb_005062_080 |
0.257125902 |
- |
- |
MLO-like protein 4 [Gossypium arboreum] |
| 3 |
Hb_000038_120 |
0.2809360998 |
- |
- |
PREDICTED: endoglucanase 9-like [Jatropha curcas] |
| 4 |
Hb_009571_010 |
0.2829704522 |
- |
- |
early-responsive to dehydration family protein [Populus trichocarpa] |
| 5 |
Hb_021576_020 |
0.2858480168 |
- |
- |
PREDICTED: uncharacterized protein LOC104426132 [Eucalyptus grandis] |
| 6 |
Hb_005725_050 |
0.2995833047 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 7 |
Hb_007919_020 |
0.3004188339 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 8 |
Hb_011930_150 |
0.306123337 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001638_180 |
0.3079063764 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 10 |
Hb_005306_010 |
0.3107066266 |
- |
- |
- |
| 11 |
Hb_003464_070 |
0.3113829852 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |
| 12 |
Hb_000010_310 |
0.3123614123 |
- |
- |
PREDICTED: probable pectate lyase 18 [Populus euphratica] |
| 13 |
Hb_002811_280 |
0.3146931395 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 14 |
Hb_000926_100 |
0.3172189265 |
- |
- |
hypothetical protein B456_007G076800 [Gossypium raimondii] |
| 15 |
Hb_003927_110 |
0.3182887246 |
- |
- |
hypothetical protein PRUPE_ppa001478mg [Prunus persica] |
| 16 |
Hb_002631_190 |
0.3209743635 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 17 |
Hb_009748_010 |
0.3220574181 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 18 |
Hb_000742_160 |
0.3243485802 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g20940 [Jatropha curcas] |
| 19 |
Hb_002998_070 |
0.3266112445 |
- |
- |
hypothetical protein CARUB_v10019139mg, partial [Capsella rubella] |
| 20 |
Hb_080128_020 |
0.329050171 |
- |
- |
conserved hypothetical protein [Ricinus communis] |