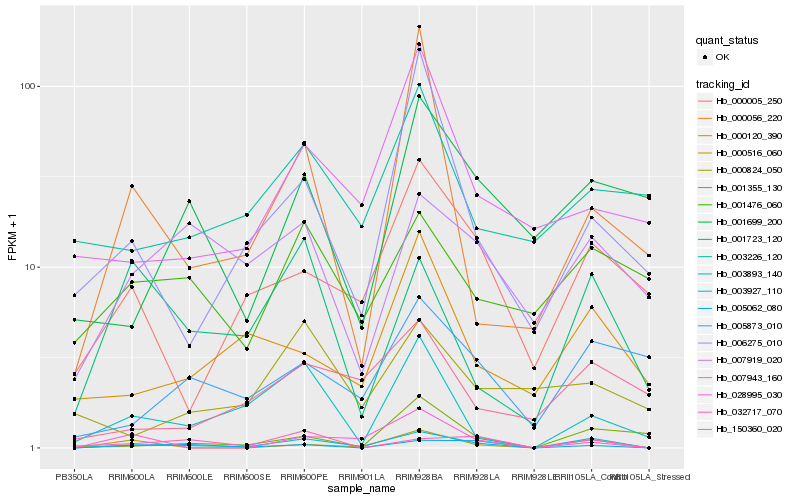
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003927_110 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa001478mg [Prunus persica] |
| 2 |
Hb_000516_060 |
0.2869005421 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 3 |
Hb_005062_080 |
0.292789571 |
- |
- |
MLO-like protein 4 [Gossypium arboreum] |
| 4 |
Hb_000005_250 |
0.3010450135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003893_140 |
0.3019277106 |
- |
- |
palmitoyl-acyl carrier protein thioesterase [Ricinus communis] |
| 6 |
Hb_005873_010 |
0.3100823301 |
- |
- |
cysteine desulfurylase, putative [Ricinus communis] |
| 7 |
Hb_000120_390 |
0.3113612325 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 8 |
Hb_001723_120 |
0.312170662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_150360_020 |
0.315736714 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 10 |
Hb_032717_070 |
0.3182887246 |
- |
- |
PREDICTED: mucin-2-like [Jatropha curcas] |
| 11 |
Hb_001355_130 |
0.3196647061 |
- |
- |
PREDICTED: uncharacterized protein LOC105137896 [Populus euphratica] |
| 12 |
Hb_007919_020 |
0.3240868315 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 13 |
Hb_006275_010 |
0.3242768877 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 14 |
Hb_028995_030 |
0.3256761803 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 15 |
Hb_001476_060 |
0.3331836144 |
- |
- |
PREDICTED: uncharacterized protein At5g01610 [Jatropha curcas] |
| 16 |
Hb_007943_160 |
0.333629744 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 17 |
Hb_001699_200 |
0.3347062194 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003226_120 |
0.3399136073 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000056_220 |
0.3442571862 |
- |
- |
PREDICTED: HVA22-like protein c [Jatropha curcas] |
| 20 |
Hb_000824_050 |
0.347517344 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |