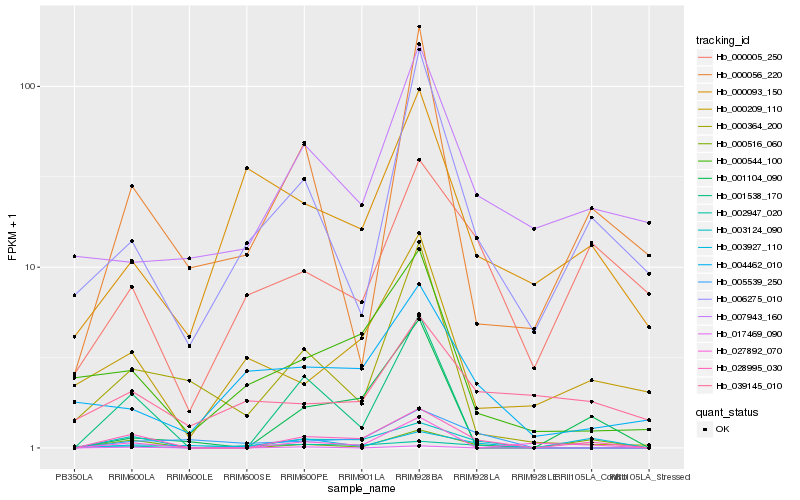
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028995_030 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 2 |
Hb_003124_090 |
0.2204948781 |
- |
- |
hypothetical protein CICLE_v10031194mg [Citrus clementina] |
| 3 |
Hb_000516_060 |
0.2276249751 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 4 |
Hb_001538_170 |
0.3024234325 |
- |
- |
- |
| 5 |
Hb_001104_090 |
0.3088983908 |
- |
- |
hypothetical protein B456_009G408400 [Gossypium raimondii] |
| 6 |
Hb_027892_070 |
0.315063792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003927_110 |
0.3256761803 |
- |
- |
hypothetical protein PRUPE_ppa001478mg [Prunus persica] |
| 8 |
Hb_006275_010 |
0.3277152471 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 9 |
Hb_000544_100 |
0.3340743764 |
- |
- |
hypothetical protein JCGZ_03636 [Jatropha curcas] |
| 10 |
Hb_000005_250 |
0.3402650407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005539_250 |
0.3436948666 |
- |
- |
- |
| 12 |
Hb_000056_220 |
0.3464991564 |
- |
- |
PREDICTED: HVA22-like protein c [Jatropha curcas] |
| 13 |
Hb_000209_110 |
0.3527282217 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 14 |
Hb_000364_200 |
0.3541721699 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_007943_160 |
0.3557449048 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 16 |
Hb_004462_010 |
0.3659802708 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 17 |
Hb_017469_090 |
0.3772145222 |
- |
- |
Integrase, catalytic region, Zinc finger, CCHC-type, Peptidase aspartic, catalytic, putative [Theobroma cacao] |
| 18 |
Hb_002947_020 |
0.3790262884 |
- |
- |
PREDICTED: uncharacterized protein LOC103330439 [Prunus mume] |
| 19 |
Hb_039145_010 |
0.379974709 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 20 |
Hb_000093_150 |
0.3805385645 |
- |
- |
6-phosphogluconolactonase, putative [Ricinus communis] |