| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
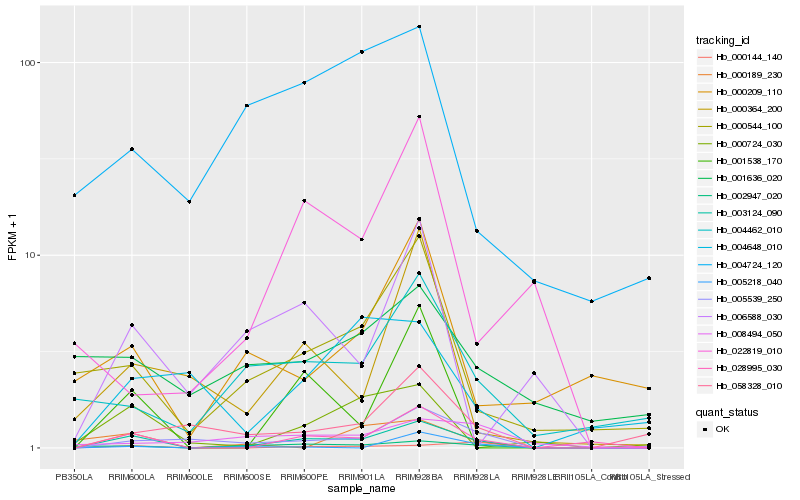
Hb_003124_090 |
0.0 |
- |
- |
hypothetical protein CICLE_v10031194mg [Citrus clementina] |
| 2 |
Hb_028995_030 |
0.2204948781 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 3 |
Hb_002947_020 |
0.2690946161 |
- |
- |
PREDICTED: uncharacterized protein LOC103330439 [Prunus mume] |
| 4 |
Hb_005539_250 |
0.2757130202 |
- |
- |
- |
| 5 |
Hb_000544_100 |
0.3085757338 |
- |
- |
hypothetical protein JCGZ_03636 [Jatropha curcas] |
| 6 |
Hb_001538_170 |
0.3203001158 |
- |
- |
- |
| 7 |
Hb_004462_010 |
0.3328476553 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 8 |
Hb_058328_010 |
0.3352633583 |
- |
- |
JHL06B08.2 [Jatropha curcas] |
| 9 |
Hb_000724_030 |
0.3438722582 |
- |
- |
PREDICTED: uncharacterized protein LOC105645165 [Jatropha curcas] |
| 10 |
Hb_000364_200 |
0.3509881548 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_006588_030 |
0.3561252165 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Jatropha curcas] |
| 12 |
Hb_000144_140 |
0.3586958524 |
- |
- |
PREDICTED: uncharacterized protein LOC105629608 [Jatropha curcas] |
| 13 |
Hb_004724_120 |
0.3638218019 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 14 |
Hb_005218_040 |
0.365257013 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 15 |
Hb_000189_230 |
0.3654772633 |
- |
- |
Auxin-induced protein [Aegilops tauschii] |
| 16 |
Hb_000209_110 |
0.3721147362 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 17 |
Hb_008494_050 |
0.3730858552 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 18 |
Hb_004648_010 |
0.3750159963 |
- |
- |
UDP-galactose/UDP-glucose transporter 4 [Morus notabilis] |
| 19 |
Hb_001636_020 |
0.3778538744 |
- |
- |
Serine/threonine-protein kinase PBS1 [Theobroma cacao] |
| 20 |
Hb_022819_010 |
0.3788796679 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |