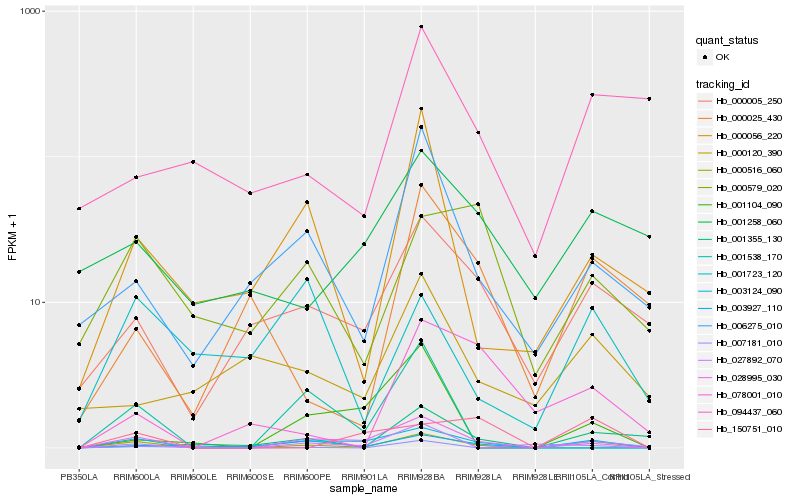
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000516_060 |
0.0 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 2 |
Hb_028995_030 |
0.2276249751 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 3 |
Hb_003927_110 |
0.2869005421 |
- |
- |
hypothetical protein PRUPE_ppa001478mg [Prunus persica] |
| 4 |
Hb_000005_250 |
0.3452111152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006275_010 |
0.3524313831 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 6 |
Hb_000056_220 |
0.359454992 |
- |
- |
PREDICTED: HVA22-like protein c [Jatropha curcas] |
| 7 |
Hb_027892_070 |
0.3693548711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007181_010 |
0.3731894889 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 9 |
Hb_003124_090 |
0.3831111375 |
- |
- |
hypothetical protein CICLE_v10031194mg [Citrus clementina] |
| 10 |
Hb_001355_130 |
0.3931823869 |
- |
- |
PREDICTED: uncharacterized protein LOC105137896 [Populus euphratica] |
| 11 |
Hb_000025_430 |
0.3953010655 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_150751_010 |
0.4011935509 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 13 |
Hb_000579_020 |
0.4024383499 |
- |
- |
hypothetical protein POPTR_0017s06850g [Populus trichocarpa] |
| 14 |
Hb_001104_090 |
0.4033494377 |
- |
- |
hypothetical protein B456_009G408400 [Gossypium raimondii] |
| 15 |
Hb_078001_010 |
0.4048922192 |
- |
- |
Uncharacterized protein TCM_000398 [Theobroma cacao] |
| 16 |
Hb_000120_390 |
0.4070976039 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 17 |
Hb_001538_170 |
0.4075928778 |
- |
- |
- |
| 18 |
Hb_001258_060 |
0.407736911 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 19 |
Hb_001723_120 |
0.408951214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_094437_060 |
0.4117108767 |
- |
- |
PREDICTED: cytochrome B5 isoform D-like [Jatropha curcas] |