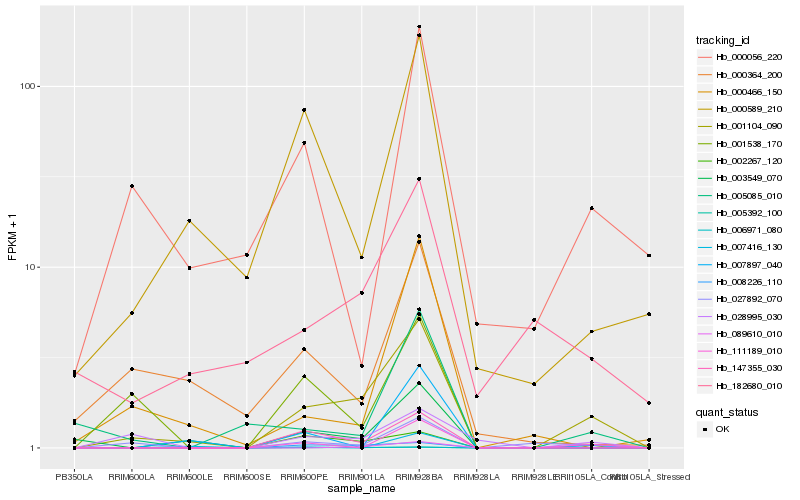
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001104_090 |
0.0 |
- |
- |
hypothetical protein B456_009G408400 [Gossypium raimondii] |
| 2 |
Hb_005392_100 |
0.2762376985 |
- |
- |
Uncharacterized protein TCM_038736 [Theobroma cacao] |
| 3 |
Hb_027892_070 |
0.2873180869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_147355_030 |
0.2979549114 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 isoform X5 [Populus euphratica] |
| 5 |
Hb_111189_010 |
0.3034066965 |
- |
- |
PREDICTED: uncharacterized protein LOC105801112 [Gossypium raimondii] |
| 6 |
Hb_028995_030 |
0.3088983908 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 7 |
Hb_008226_110 |
0.3136136879 |
- |
- |
uncharacterized protein LOC100501343 [Zea mays] |
| 8 |
Hb_003549_070 |
0.3161837678 |
- |
- |
- |
| 9 |
Hb_000589_210 |
0.316480469 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 10 |
Hb_182680_010 |
0.3190887429 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 11 |
Hb_007897_040 |
0.3321707835 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_001538_170 |
0.3336304019 |
- |
- |
- |
| 13 |
Hb_006971_080 |
0.3350953481 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 14 |
Hb_005085_010 |
0.3361276465 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 15 |
Hb_000364_200 |
0.3372779613 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_002267_120 |
0.3390084664 |
- |
- |
hypothetical protein B456_009G136300 [Gossypium raimondii] |
| 17 |
Hb_000056_220 |
0.3394736482 |
- |
- |
PREDICTED: HVA22-like protein c [Jatropha curcas] |
| 18 |
Hb_007416_130 |
0.3400458534 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 19 |
Hb_000466_150 |
0.3404866608 |
- |
- |
hypothetical protein JCGZ_02123 [Jatropha curcas] |
| 20 |
Hb_089610_010 |
0.3436776786 |
- |
- |
PREDICTED: uncharacterized protein LOC105785094 [Gossypium raimondii] |